5BYC
 
 | | Crystal structure of human ribokinase in C2 spacegroup | | Descriptor: | Ribokinase, SODIUM ION | | Authors: | Park, J, Chakrabarti, J, Singh, B, Gupta, R.S, Junop, M.S. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of unliganded human ribokinase in C2 spacegroup
To Be Published
|
|
5XYK
 
 | | Structure of Transferase | | Descriptor: | ARGININE, MANGANESE (II) ION, Putative cytoplasmic protein, ... | | Authors: | Park, J.B, Yoo, Y, Kim, J, Cho, H.S. | | Deposit date: | 2017-07-09 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of Transferase
To Be Published
|
|
5CTT
 
 | |
5CTR
 
 | |
5CTQ
 
 | |
4R4S
 
 | | Crystal structure of chimeric beta-lactamase cTEM-19m at 1.1 angstrom resolution | | Descriptor: | Beta-lactamase TEM,Beta-lactamase PSE-4, CHLORIDE ION, MAGNESIUM ION | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-08-19 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Structural Dynamics of Engineered beta-Lactamases Vary Broadly on Three Timescales yet Sustain Native Function.
Sci Rep, 9, 2019
|
|
7CZ3
 
 | |
6AI4
 
 | |
5TUF
 
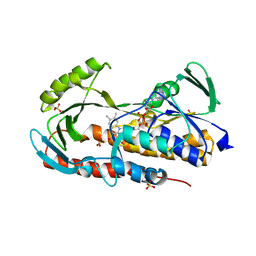 | | Crystal structure of tetracycline destructase Tet(50) in complex with anhydrotetracycline | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUK
 
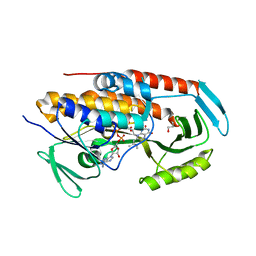 | | Crystal structure of tetracycline destructase Tet(51) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Tetracycline destructase Tet(51) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUI
 
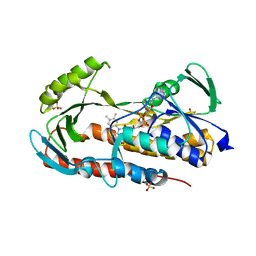 | | Crystal structure of tetracycline destructase Tet(50) in complex with chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUE
 
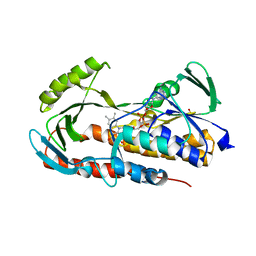 | | Crystal structure of tetracycline destructase Tet(50) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Tetracycline destructase Tet(50) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
4R4R
 
 | | Crystal structure of chimeric beta-lactamase cTEM-19m at 1.2 angstrom resolution | | Descriptor: | Beta-lactamase TEM,Beta-lactamase PSE-4, CHLORIDE ION, MAGNESIUM ION | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-08-19 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Structural Dynamics of Engineered beta-Lactamases Vary Broadly on Three Timescales yet Sustain Native Function.
Sci Rep, 9, 2019
|
|
5TUM
 
 | | Crystal structure of tetracycline destructase Tet(56) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tetracycline destructase Tet(56) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUL
 
 | |
7XQW
 
 | | Formate dehydrogenase (FDH) from Methylobacterium extorquens AM1 (MeFDH1) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Park, J, Heo, Y.Y, Roh, S.H, Lee, H.H. | | Deposit date: | 2022-05-09 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Enzymatic conversion of CO2 in real flue gas to molar-scale formate
To Be Published
|
|
5Z0B
 
 | |
6KBM
 
 | | Crystal structure of Vac8 bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
4QY6
 
 | | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations | | Descriptor: | Beta-lactamase TEM, Beta-lactamase PSE-4, CHLORIDE ION, ... | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations
To be Published
|
|
6KBN
 
 | | Crystal structure of Vac8 (del 19-33) bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
6JQQ
 
 | | KatE H392C from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, J.B, Cho, H.-S. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | KatE H392C from Escherichia coli
To Be Published
|
|
4OF1
 
 | |
4NFK
 
 | | Crystal structure of human FPPS in complex with nickel, JDS05120, and sulfate | | Descriptor: | Farnesyl pyrophosphate synthase, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Park, J, De schutter, J.W, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic and thermodynamic characterization of phenylaminopyridine bisphosphonates binding to human farnesyl pyrophosphate synthase.
PLoS ONE, 12, 2017
|
|
4QXS
 
 | | Crystal structure of human FPPS in complex with WC01088 | | Descriptor: | (2-{2-[(2S)-3-methylbutan-2-yl]-5-phenyl-1H-indol-3-yl}ethane-1,1-diyl)bis(phosphonic acid), Farnesyl pyrophosphate synthase, GLYCEROL, ... | | Authors: | Park, J, Zielinski, M, Weiling, C, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the molecular and structural elements of ligands binding to the active site versus an allosteric pocket of the human farnesyl pyrophosphate synthase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4QY5
 
 | | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations | | Descriptor: | Beta-lactamase TEM,Beta-lactamase PSE-4, CHLORIDE ION, MAGNESIUM ION | | Authors: | Park, J, Gobeil, S, Pelletier, J.N, Berghuis, A.M. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structures of chimeric beta-lactamase cTEM-19m showing different conformations
To be Published
|
|
