2DS5
 
 | | Structure of the ZBD in the orthorhomibic crystal from | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Song, H.K, Park, E.Y, Lee, B.G, Hong, S.B. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
1KOZ
 
 | | SOLUTION STRUCTURE OF OMEGA-GRAMMOTOXIN SIA | | Descriptor: | Voltage-dependent Channel Inhibitor | | Authors: | Takeuchi, K, Park, E.J, Lee, C.W, Kim, J.I, Takahashi, H, Swartz, K.J, Shimada, I. | | Deposit date: | 2001-12-25 | | Release date: | 2002-08-28 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega-grammotoxin SIA, a gating modifier of P/Q and N-type Ca(2+) channel.
J.Mol.Biol., 321, 2002
|
|
5EUL
 
 | | Structure of the SecA-SecY complex with a translocating polypeptide substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AYC08, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Li, L, Park, E, Ling, J, Ingram, J, Ploegh, H, Rapoport, T.A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of a substrate-engaged SecY protein-translocation channel.
Nature, 531, 2016
|
|
7WLG
 
 | | Cryo-EM structure of GH31 alpha-1,3-glucosidase from Lactococcus lactis subsp. cremoris | | Descriptor: | Alpha-xylosidase | | Authors: | Ikegaya, M, Moriya, T, Adachi, N, Kawasaki, M, Park, E.Y, Miyazaki, T. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7KAO
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAM
 
 | | Cryo-EM structure of the Sec complex from T. lanuginosus, wild-type, class with Sec62, plug-closed conformation | | Descriptor: | Protein transport channel Sec61 complex, alpha subunit (Sec61), beta subunit (Sbh1), ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAN
 
 | | Cryo-EM structure of the Sec complex from T. lanuginosus, Sec62-lacking mutant (Delta Sec62) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Protein transport channel Sec61 complex, alpha subunit (Sec61), ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAQ
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class with Sec62, conformation 2 (C2) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAJ
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class with Sec62, conformation 2 (C2) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAS
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec63 FN3 mutant, class with Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAL
 
 | | Cryo-EM structure of the Sec complex from T. lanuginosus, wild-type, class with Sec62, plug-open conformation | | Descriptor: | Protein transport channel Sec61 complex, alpha subunit (Sec61), beta subunit (Sbh1), ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAH
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAT
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore ring and Sec63 FN3 double mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1R6L
 
 | | Crystal Structure Of The tRNA Processing Enzyme Rnase pH From Pseudomonas Aeruginosa | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Ribonuclease PH, SULFATE ION | | Authors: | Choi, J.M, Park, E.Y, Kim, J.H, Chang, S.K, Cho, Y. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the functional importance of the hexameric ring structure of RNase PH
J.BIOL.CHEM., 279, 2004
|
|
7KAI
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class with Sec62, conformation 1 (C1) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAR
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec63 FN3 mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAU
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore ring and Sec63 FN3 double mutant, class with Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAP
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class with Sec62, conformation 1 (C1) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KB5
 
 | |
7M0Y
 
 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and Trametinib | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M0Z
 
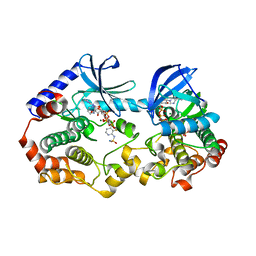 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and CH5126766 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, N-(3-fluoro-4-{[4-methyl-2-oxo-7-(pyrimidin-2-yloxy)-2H-chromen-3-yl]methyl}pyridin-2-yl)-N'-methylsulfuric diamide, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M0V
 
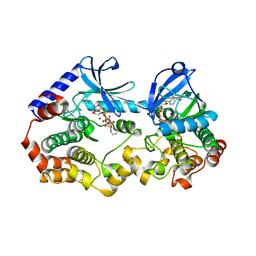 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and Cobimetinib | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M0W
 
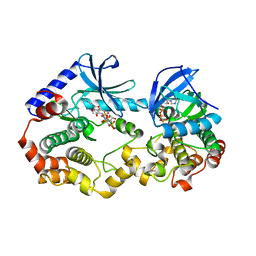 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and Pimasertib | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, N-[(2S)-2,3-dihydroxypropyl]-3-[(2-fluoro-4-iodophenyl)amino]pyridine-4-carboxamide, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M0X
 
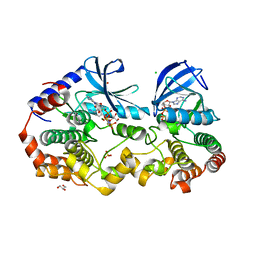 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and PD0325901 | | Descriptor: | CHLORIDE ION, Dual specificity mitogen-activated protein kinase kinase 1, GLYCEROL, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M0U
 
 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and Binimetinib | | Descriptor: | 5-[(4-bromo-2-fluorophenyl)amino]-4-fluoro-N-(2-hydroxyethoxy)-1-methyl-1H-benzimidazole-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
