2LAF
 
 | |
2LAE
 
 | |
3OG5
 
 | | Crystal Structure of BamA POTRA45 tandem | | Descriptor: | Outer membrane protein assembly complex, YaeT protein | | Authors: | Gatzeva-Topalova, P.Z, Warner, L.R, Pardi, A, Sousa, M.C. | | Deposit date: | 2010-08-16 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure and Flexibility of the Complete Periplasmic Domain of BamA: The Protein Insertion Machine of the Outer Membrane
Structure, 18, 2010
|
|
1EHT
 
 | |
1RNG
 
 | |
1BNB
 
 | |
1NBR
 
 | |
1LDZ
 
 | |
1AQO
 
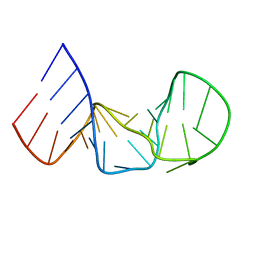 | | IRON RESPONSIVE ELEMENT RNA HAIRPIN, NMR, 15 STRUCTURES | | Descriptor: | IRON RESPONSIVE ELEMENT RNA HAIRPIN | | Authors: | Addess, K.J, Pardi, A. | | Deposit date: | 1997-07-31 | | Release date: | 1998-02-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory binding proteins.
J.Mol.Biol., 274, 1997
|
|
1ZIG
 
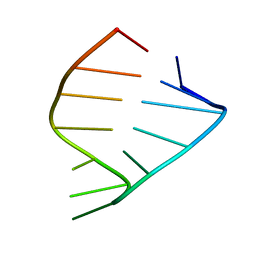 | | GAGA RNA TETRALOOP, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*GP*AP*GP*AP*GP*CP*CP*U)-3') | | Authors: | Jucker, F.M, Heus, H.A, Yip, P.F, Moors, E, Pardi, A. | | Deposit date: | 1996-07-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A network of heterogeneous hydrogen bonds in GNRA tetraloops.
J.Mol.Biol., 264, 1996
|
|
1ZIH
 
 | | GCAA RNA TETRALOOP, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*GP*CP*AP*AP*GP*CP*CP*U)-3') | | Authors: | Jucker, F.M, Heus, H.A, Yip, P.F, Moors, E, Pardi, A. | | Deposit date: | 1996-07-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A network of heterogeneous hydrogen bonds in GNRA tetraloops.
J.Mol.Biol., 264, 1996
|
|
1ZIF
 
 | | GAAA RNA TETRALOOP, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*GP*AP*AP*AP*GP*CP*CP*U)-3') | | Authors: | Jucker, F.M, Heus, H.A, Yip, P.F, Moors, E, Pardi, A. | | Deposit date: | 1996-07-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A network of heterogeneous hydrogen bonds in GNRA tetraloops.
J.Mol.Biol., 264, 1996
|
|
2LDZ
 
 | |
2K4C
 
 | | tRNAPhe-based homology model for tRNAVal refined against base N-H RDCs in two media and SAXS data | | Descriptor: | 76-MER | | Authors: | Grishaev, A, Ying, J, Canny, M.D, Pardi, A, Bax, A. | | Deposit date: | 2008-06-04 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data.
J.Biomol.Nmr, 42, 2008
|
|
283D
 
 | | A CURVED RNA HELIX INCORPORATING AN INTERNAL LOOP WITH G-A AND A-A NON-WATSON-CRICK BASE PAIRING | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*GP*GP*CP*CP*GP*AP*AP*AP*GP*GP*CP*C)-3') | | Authors: | Baeyens, K.J, De Bondt, H.L, Pardi, A, Holbrook, S.R. | | Deposit date: | 1996-09-03 | | Release date: | 1996-09-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A curved RNA helix incorporating an internal loop with G.A and A.A non-Watson-Crick base pairing.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
