1GZX
 
 | | Oxy T State Haemoglobin - Oxygen bound at all four haems | | 分子名称: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | 著者 | Paoli, M, Liddington, R, Tame, J, Wilkinson, A, Dodson, G. | | 登録日 | 2002-06-07 | | 公開日 | 2002-07-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of T State Haemoglobin with Oxygen Bound at All Four Haems.
J.Mol.Biol., 256, 1996
|
|
1QJS
 
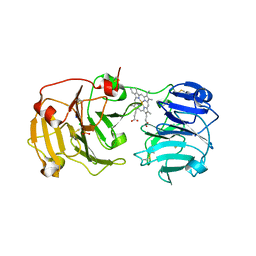 | | mammalian blood serum haemopexin glycosylated-native protein and in complex with its ligand haem | | 分子名称: | CHLORIDE ION, HEMOPEXIN, PHOSPHATE ION, ... | | 著者 | Paoli, M, Baker, H.M, Morgan, W.T, Smith, A, Baker, E.N. | | 登録日 | 1999-07-01 | | 公開日 | 2000-02-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of Hemopexin Reveals a Novel High-Affinity Heme Site Formed between Two Beta-Propeller Domains.
Nat.Struct.Biol., 6, 1999
|
|
1QHU
 
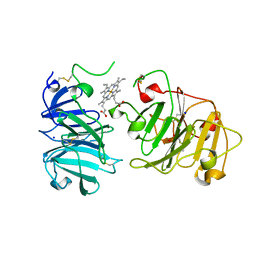 | | MAMMALIAN BLOOD SERUM HAEMOPEXIN DEGLYCOSYLATED AND IN COMPLEX WITH ITS LIGAND HAEM | | 分子名称: | CHLORIDE ION, PHOSPHATE ION, PROTEIN (HEMOPEXIN), ... | | 著者 | Paoli, M, Baker, H.M, Morgan, W.T, Smith, A, Baker, E.N. | | 登録日 | 1999-05-27 | | 公開日 | 1999-10-06 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two beta-propeller domains.
Nat.Struct.Biol., 6, 1999
|
|
8OMT
 
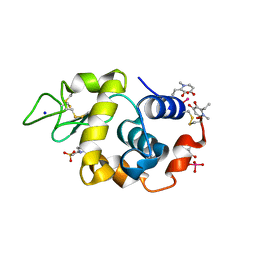 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(empp)2] (Structure C) | | 分子名称: | 1-methyl-2-ethyl-3-hydroxy-4(1H)-pyridinone)V(IV)O4, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme C, ... | | 著者 | Paolillo, M, Merlino, A, Ferraro, G. | | 登録日 | 2023-03-31 | | 公開日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (1.097 Å) | | 主引用文献 | Implications of Protein Interaction in the Speciation of Potential V IV O-Pyridinone Drugs.
Inorg.Chem., 62, 2023
|
|
8OM8
 
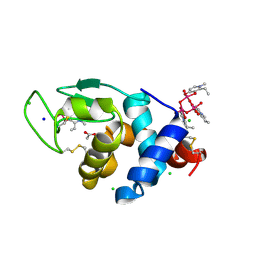 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(empp)2] (Structure A) | | 分子名称: | 1-methyl-2-ethyl-3-hydroxy-4(1H)-pyridinone)V(IV)O4, ACETATE ION, CHLORIDE ION, ... | | 著者 | Paolillo, M, Ferraro, G, Merlino, A. | | 登録日 | 2023-03-31 | | 公開日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Implications of Protein Interaction in the Speciation of Potential V IV O-Pyridinone Drugs.
Inorg.Chem., 62, 2023
|
|
8OMS
 
 | |
8AJ4
 
 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure A') | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8,8-bis($l^{1}-oxidanyl)-2,2'-dimethyl-8,8'-spirobi[3$l^{4},7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1(6),2,4-triene], Lysozyme, ... | | 著者 | Paolillo, M, Merlino, A, Ferraro, G. | | 登録日 | 2022-07-27 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
8AJ3
 
 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure A) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8,8,8,8-tetrakis($l^{1}-oxidanyl)-2-methyl-3,7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1,5-diene, 8,8-bis($l^{1}-oxidanyl)-2,2'-dimethyl-8,8'-spirobi[3$l^{4},7,9-trioxa-8$l^{6}-vanadabicyclo[4.3.0]nona-1(6),2,4-triene], ... | | 著者 | Paolillo, M, Merlino, A, Ferraro, G. | | 登録日 | 2022-07-27 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
8AJ5
 
 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(malt)2] (Structure B) | | 分子名称: | ACETATE ION, Lysozyme, NITRATE ION, ... | | 著者 | Paolillo, M, Merlino, A, Ferraro, G. | | 登録日 | 2022-07-27 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Multiple and Variable Binding of Pharmacologically Active Bis(maltolato)oxidovanadium(IV) to Lysozyme.
Inorg.Chem., 61, 2022
|
|
2V79
 
 | | Crystal Structure of the N-terminal domain of DnaD from Bacillus Subtilis | | 分子名称: | CHLORIDE ION, DNA REPLICATION PROTEIN DNAD, SODIUM ION | | 著者 | Schneider, S, Zhang, W, Soultanas, P, Paoli, M. | | 登録日 | 2007-07-27 | | 公開日 | 2008-01-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the N-Terminal Oligomerization Domain of Dnad Reveals a Unique Tetramerization Motif and Provides Insights Into Scaffold Formation.
J.Mol.Biol., 376, 2008
|
|
1U36
 
 | | Crystal structure of WLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-21 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3Y
 
 | | Crystal structure of ILAC mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-22 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U42
 
 | | Crystal structure of MLAM mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-23 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.699 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U41
 
 | | Crystal structure of YLGV mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-23 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3Z
 
 | | Crystal structure of MLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-23 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3J
 
 | | Crystal structure of MLAV mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-22 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
3EIF
 
 | | 1.9 angstrom crystal structure of the active form of the C5a peptidase from Streptococcus pyogenes (ScpA) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | 著者 | Cooney, J.C, Kagawa, T.F, O'Connell, M.R, Paoli, M, Mouat, P, O'Toole, P.W. | | 登録日 | 2008-09-15 | | 公開日 | 2009-02-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Model for Substrate Interactions in C5a Peptidase from Streptococcus pyogenes: A 1.9 A Crystal Structure of the Active Form of ScpA
J.Mol.Biol., 386, 2009
|
|
2BZS
 
 | | Binding of anti-cancer prodrug CB1954 to the activating enzyme NQO2 revealed by the crystal structure of their complex. | | 分子名称: | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, NRH DEHYDROGENASE [QUINONE] 2, ... | | 著者 | Abu Khader, M.M, Heap, J.T, De Matteis, C, Kellam, B, Doughty, S.W, Minton, N, Paoli, M. | | 登録日 | 2005-08-22 | | 公開日 | 2005-09-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Binding of the Anticancer Prodrug Cb1954 to the Activating Enzyme Nqo2 Revealed by the Crystal Structure of Their Complex.
J.Med.Chem., 48, 2005
|
|
3VC0
 
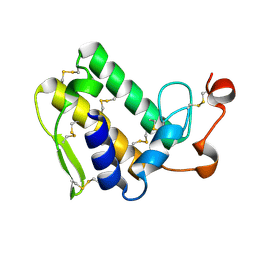 | | Crystal structure of Taipoxin beta subunit isoform 1 | | 分子名称: | Phospholipase A2 homolog, taipoxin beta chain | | 著者 | Cendron, L, Micetic, I, Polverino de Laureto, P, Beltramini, M, Paoli, M. | | 登録日 | 2012-01-03 | | 公開日 | 2012-07-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural analysis of trimeric phospholipase A(2) neurotoxin from the Australian taipan snake venom.
Febs J., 279, 2012
|
|
3VBZ
 
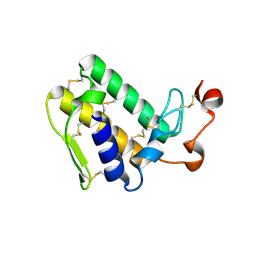 | | Crystal structure of Taipoxin beta subunit isoform 2 | | 分子名称: | Phospholipase A2 homolog, taipoxin beta chain | | 著者 | Cendron, L, Micetic, I, Polverino, P, Beltramini, M, Paoli, M. | | 登録日 | 2012-01-03 | | 公開日 | 2012-07-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural analysis of trimeric phospholipase A(2) neurotoxin from the Australian taipan snake venom.
Febs J., 279, 2012
|
|
2J0P
 
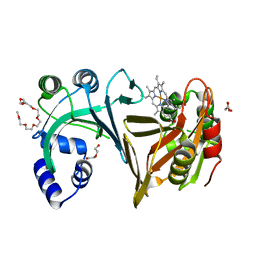 | | Structure of the haem-chaperone Proteobacteria-protein HemS | | 分子名称: | DI(HYDROXYETHYL)ETHER, DODECAETHYLENE GLYCOL, HEMIN TRANSPORT PROTEIN HEMS, ... | | 著者 | Schneider, S, Sharp, K.H, Barker, P.D, Paoli, M. | | 登録日 | 2006-08-04 | | 公開日 | 2006-08-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | An Induced Fit Conformational Change Underlies the Binding Mechanism of the Heme Transport Proteobacteria-Protein Hems.
J.Biol.Chem., 281, 2006
|
|
2J0R
 
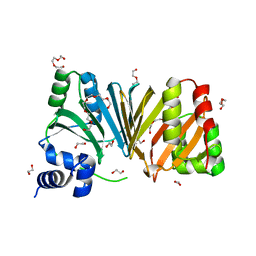 | | Structure of the haem-chaperone Proteobacteria-protein HemS | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DODECAETHYLENE GLYCOL, ... | | 著者 | Schneider, S, Sharp, K.H, Barker, P.D, Paoli, M. | | 登録日 | 2006-08-04 | | 公開日 | 2006-08-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | An Induced Fit Conformational Change Underlies the Binding Mechanism of the Heme Transport Proteobacteria-Protein Hems.
J.Biol.Chem., 281, 2006
|
|
2O6P
 
 | | Crystal Structure of the heme-IsdC complex | | 分子名称: | CHLORIDE ION, Iron-regulated surface determinant protein C, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Sharp, K.H, Schneider, S, Cockayne, A, Paoli, M. | | 登録日 | 2006-12-08 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus.
J. Biol. Chem., 282, 2007
|
|
