3V1S
 
 | | Scaffold tailoring by a newly detected Pictet-Spenglerase ac-tivity of strictosidine synthase (STR1): from the common tryp-toline skeleton to the rare piperazino-indole framework | | Descriptor: | 2-(1H-indol-1-yl)ethanamine, Strictosidine synthase | | Authors: | Wu, F, Zhu, H, Sun, L, Rajendran, C, Wang, M, Ren, X, Panjikar, S, Cherkasov, A, Zou, H, Stoeckigt, J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Scaffold Tailoring by a Newly Detected Pictet-Spenglerase Activity of Strictosidine Synthase: From the Common Tryptoline Skeleton to the Rare Piperazino-indole Framework
J.Am.Chem.Soc., 134, 2012
|
|
8TX9
 
 | |
3V0T
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine Reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.333 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3V0U
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3V0S
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3UYI
 
 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3U5U
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3ZJ7
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-1 | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
4BEB
 
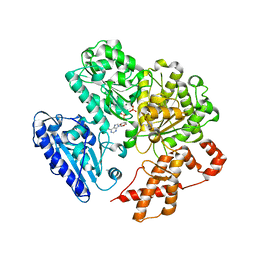 | | MUTANT (K220E) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
4BEC
 
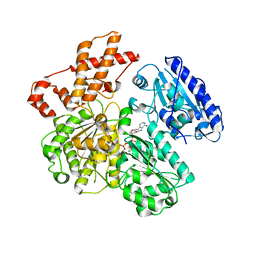 | | MUTANT (K220A) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
4BE7
 
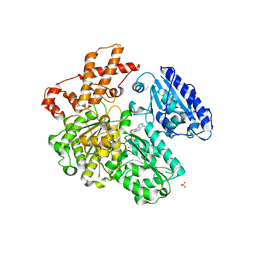 | | MUTANT (K220R) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-06 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
4BAJ
 
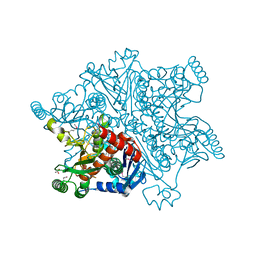 | | MYCOBACTERIUM TUBERCULOSIS CHORISMATE SYNTHASE after exposure to 266nm UV laser | | Descriptor: | ACETATE ION, CHORISMATE SYNTHASE | | Authors: | Pereira, P.J.B, Royant, A, Panjikar, S, de Sanctis, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In-house UV radiation-damage-induced phasing of selenomethionine-labeled protein structures.
J. Struct. Biol., 181, 2013
|
|
4B4P
 
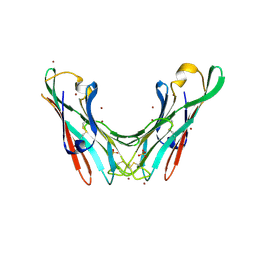 | | Crystal Structure of the lectin domain of F18 fimbrial adhesin FedF. | | Descriptor: | BROMIDE ION, F18 FIMBRIAL ADHESIN AC, SULFATE ION | | Authors: | Moonens, K, Bouckaert, J, Coddens, A, Tran, T, Panjikar, S, De Kerpel, M, Cox, E, Remaut, H, De Greve, H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight in Histo-Blood Group Binding by the F18 Fimbrial Adhesin Fedf.
Mol.Microbiol., 86, 2012
|
|
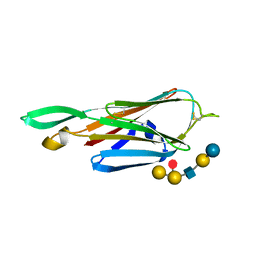
4A3Y
 
 | | Crystal structure of Raucaffricine glucosidase from ajmaline biosynthesis pathway | | Descriptor: | GLYCEROL, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Barleben, L, Rajendran, C, Lin, H, Stoeckigt, J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity.
Acs Chem.Biol., 7, 2012
|
|
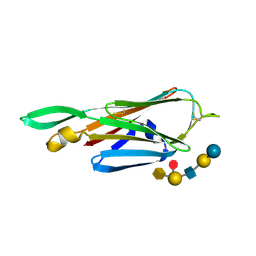
4B4R
 
 | | Crystal Structure of the lectin domain of F18 fimbrial adhesin FedF in complex with blood group B type 1 hexasaccharide | | Descriptor: | F18 FIMBRIAL ADHESIN AC, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Bouckaert, J, Coddens, A, Tran, T, Panjikar, S, De Kerpel, M, Cox, E, Remaut, H, De Greve, H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight in Histo-Blood Group Binding by the F18 Fimbrial Adhesin Fedf.
Mol.Microbiol., 86, 2012
|
|
4B4Q
 
 | | Crystal Structure of the lectin domain of F18 fimbrial adhesin FedF in complex with blood group A type 1 hexasaccharide | | Descriptor: | F18 FIMBRIAL ADHESIN AC, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Bouckaert, J, Coddens, A, Tran, T, Panjikar, S, De Kerpel, M, Cox, E, Remaut, H, De Greve, H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight in Histo-Blood Group Binding by the F18 Fimbrial Adhesin Fedf.
Mol.Microbiol., 86, 2012
|
|
4BAI
 
 | | Mycobacterium tuberculosis Chorismate synthase before exposure to 266 nm UV laser | | Descriptor: | ACETATE ION, CHORISMATE SYNTHASE | | Authors: | Pereira, P.J.B, Royant, A, Panjikar, S, de Sanctis, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In-house UV radiation-damage-induced phasing of selenomethionine-labeled protein structures.
J. Struct. Biol., 181, 2013
|
|
2HHC
 
 | | Crystal structure of fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-06-28 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
2HLH
 
 | | Crystal structure of fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | Nodulation fucosyltransferase, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
2J6G
 
 | | FaeG from F4ac ETEC strain 5_95, produced in tobacco plant chloroplast | | Descriptor: | ACETATE ION, FAEG | | Authors: | Van Molle, I, Joensuu, J.J, Buts, L, Panjikar, S, Kotiaho, M, Bouckaert, J, Wyns, L, Niklander-Teeri, V, De Greve, H. | | Deposit date: | 2006-09-28 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Chloroplasts Assemble the Major Subunit Faeg of Escherichia Coli F4 (K88) Fimbriae Into Strand-Swapped Dimers
J.Mol.Biol., 368, 2007
|
|
2J6R
 
 | | FaeG from F4ac ETEC strain GIS26, produced in tobacco plant chloroplast | | Descriptor: | K88 FIMBRIAL PROTEIN, PHOSPHATE ION | | Authors: | Van Molle, I, Joensuu, J.J, Buts, L, Panjikar, S, Kotiaho, M, Bouckaert, J, Wyns, L, Niklander-Teeri, V, De Greve, H. | | Deposit date: | 2006-10-03 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chloroplasts Assemble the Major Subunit Faeg of Escherichia Coli F4 (K88) Fimbriae Into Strand-Swapped Dimers
J.Mol.Biol., 368, 2007
|
|
2JF6
 
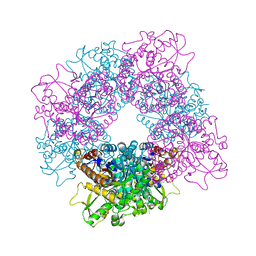 | | Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine | | Descriptor: | METHYL (2S,3R,4S)-3-ETHYL-2-(BETA-D-GLUCOPYRANOSYLOXY)-4-[(1S)-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLIN-1-YLMETHYL]-3,4-DIHYDRO-2H-PYRAN-5-CARBOXYLATE, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Barleben, L, Panjikar, S, Ruppert, M, Koepke, J, Stockigt, J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Molecular Architecture of Strictosidine Glucosidase - the Gateway to the Biosynthesis of the Monoterpenoid Indole Alkaloid Family
Plant Cell, 19, 2007
|
|
2JCG
 
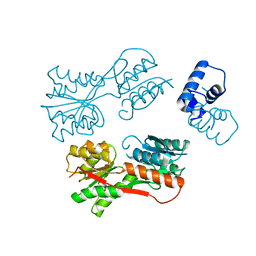 | | Apo form of the catabolite control protein A (ccpA) from bacillus megaterium, with the DNA binding domain | | Descriptor: | CALCIUM ION, GLUCOSE-RESISTANCE AMYLASE REGULATOR | | Authors: | Singh, R.K, Panjikar, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2006-12-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Apo Form of the Catabolite Control Protein a (Ccpa) from Bacillus Megaterium with a DNA-Binding Domain.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2JF7
 
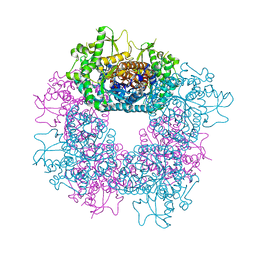 | | Structure of Strictosidine Glucosidase | | Descriptor: | STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Barleben, L, Panjikar, S, Ruppert, M, Koepke, J, Stockigt, J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Molecular Architecture of Strictosidine Glucosidase - the Gateway to the Biosynthesis of the Monoterpenoid Indole Alkaloid Family
Plant Cell, 19, 2007
|
|
2JKG
 
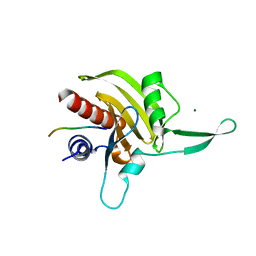 | | Plasmodium falciparum profilin | | Descriptor: | MAGNESIUM ION, OCTAPROLINE PEPTIDE, PROFILIN | | Authors: | Kursula, I, Kursula, P, Ganter, M, Panjikar, S, Matuschewski, K, Schueler, H. | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis for Parasite-Specific Functions of the Divergent Profilin of Plasmodium Falciparum.
Structure, 16, 2008
|
|
