8IJD
 
 | | Cryo-EM structure of human HCAR2-Gi complex with MK-6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJ3
 
 | | Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-24 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJB
 
 | | Cryo-EM structure of human HCAR2-Gi complex with acipimox | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
7WFD
 
 | | Left PSI in the cyclic electron transport supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pan, X, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
3JUE
 
 | | Crystal Structure of ArfGAP and ANK repeat domain of ACAP1 | | Descriptor: | ARFGAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | Authors: | Pang, X, Zhang, K, Ma, J, Zhou, Q, Sun, F. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
4NSW
 
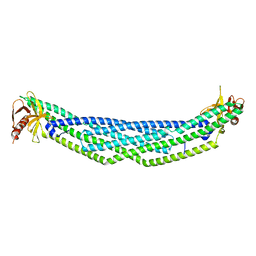 | | Crystal structure of the BAR-PH domain of ACAP1 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | | Authors: | Pang, X, Zhang, K, Ma, J, Zhou, Q, Sun, F. | | Deposit date: | 2013-11-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A PH Domain in ACAP1 Possesses Key Features of the BAR Domain in Promoting Membrane Curvature
Dev.Cell, 31, 2014
|
|
1DJM
 
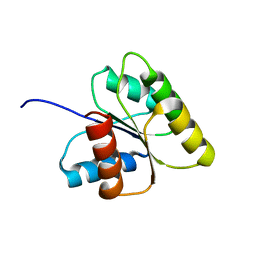 | | SOLUTION STRUCTURE OF BEF3-ACTIVATED CHEY FROM ESCHERICHIA COLI | | Descriptor: | CHEMOTAXIS PROTEIN Y | | Authors: | Cho, H.S, Lee, S.Y, Yan, D, Pan, X, Parkinson, J.S, Kustu, S, Wemmer, D.E, Pelton, J.G. | | Deposit date: | 1999-12-03 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of activated CheY.
J.Mol.Biol., 297, 2000
|
|
3QQ4
 
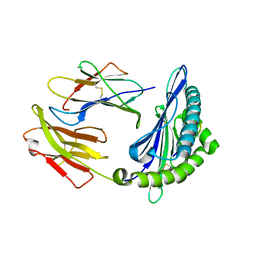 | | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, VP35 | | Authors: | Zhang, N, Qi, J, Gao, F, Pan, X, Chen, R, Li, Q, Chen, Z, Li, X, Xia, C, Gao, G.F. | | Deposit date: | 2011-02-15 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides.
J.Virol., 85, 2011
|
|
3QQ3
 
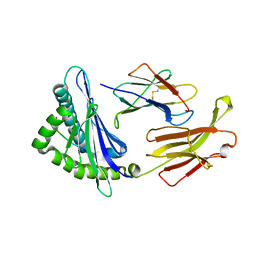 | | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides | | Descriptor: | 9-mer peptide from Neuraminidase, Beta-2-microglobulin, MHC class I antigen | | Authors: | Zhang, N, Qi, J, Gao, F, Pan, X, Chen, R, Li, Q, Chen, Z, Li, X, Xia, C, Gao, G.F. | | Deposit date: | 2011-02-15 | | Release date: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides.
J.Virol., 85, 2011
|
|
6PE0
 
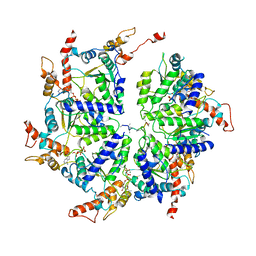 | | Msp1 (E214Q)-substrate complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Membrane-spanning ATPase-like protein, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6PDY
 
 | | Msp1-substrate complex in open conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6PDW
 
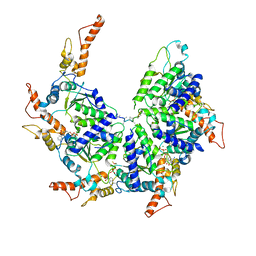 | | Msp1-substrate complex in closed conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6W90
 
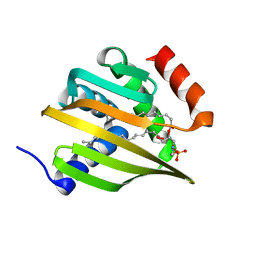 | | De novo designed NTF2 fold protein NT-9 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, NTF2 fold protein loop-helix-loop design NT-9 | | Authors: | Thompson, M.C, Pan, X, Liu, L, Fraser, J.S, Kortemme, T. | | Deposit date: | 2020-03-21 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Expanding the space of protein geometries by computational design of de novo fold families.
Science, 369, 2020
|
|
6LQA
 
 | | voltage-gated sodium channel Nav1.5 with quinidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Quinidine, Sodium channel protein type 5 subunit alpha | | Authors: | Yan, N, Li, Z, Pan, X, Huang, G. | | Deposit date: | 2020-01-13 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for Pore Blockade of the Human Cardiac Sodium Channel Na v 1.5 by the Antiarrhythmic Drug Quinidine*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4IX3
 
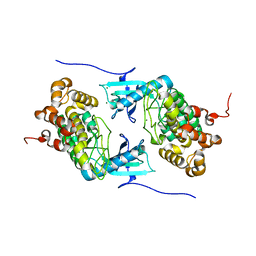 | | Crystal structure of a Stt7 homolog from Micromonas algae | | Descriptor: | MAGNESIUM ION, MsStt7d protein | | Authors: | Guo, J, Wei, X, Li, M, Pan, X, Chang, W, Liu, Z. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the catalytic domain of a state transition kinase homolog from Micromonas algae
Protein Cell, 4, 2013
|
|
4IX5
 
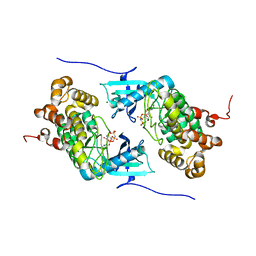 | | Crystal structure of a Stt7 homolog from Micromonas algae in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, MsStt7d protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Guo, J, Wei, X, Li, M, Pan, X, Chang, W, Liu, Z. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the catalytic domain of a state transition kinase homolog from Micromonas algae
Protein Cell, 4, 2013
|
|
4IX6
 
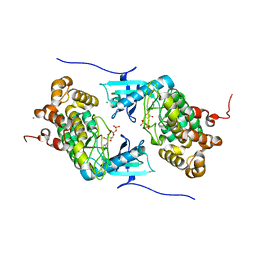 | | Crystal structure of a Stt7 homolog from Micromonas algae soaked with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MsStt7d protein | | Authors: | Guo, J, Wei, X, Li, M, Pan, X, Chang, W, Liu, Z. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the catalytic domain of a state transition kinase homolog from Micromonas algae
Protein Cell, 4, 2013
|
|
4IX4
 
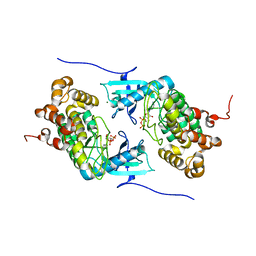 | | Crystal structure of a Stt7 homolog from Micromonas algae in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MsStt7d protein | | Authors: | Guo, J, Wei, X, Li, M, Pan, X, Chang, W, Liu, Z. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure of the catalytic domain of a state transition kinase homolog from Micromonas algae
Protein Cell, 4, 2013
|
|
1YU9
 
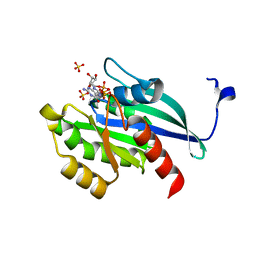 | | GppNHp-Bound Rab4A | | Descriptor: | GTP-binding protein, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-13 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1Z08
 
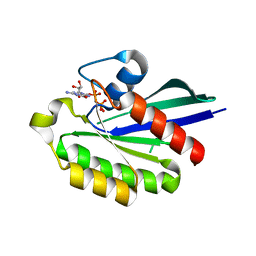 | | GppNHp-Bound Rab21 Q53G mutant GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-21 | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-01 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1YZT
 
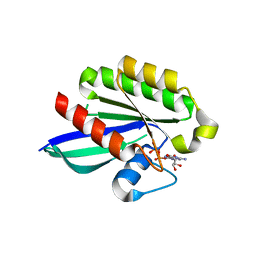 | | GppNHp-Bound Rab21 GTPase at 2.05 A Resolution | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-21 | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1YVD
 
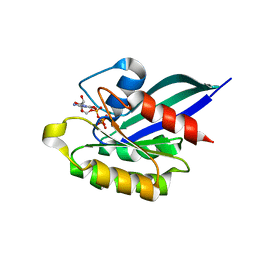 | | GppNHp-Bound Rab22 GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-22A | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-02-15 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1Z0K
 
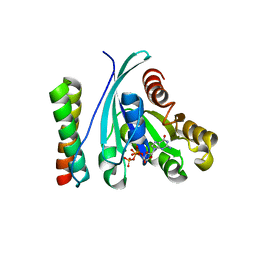 | | Structure of GTP-Bound Rab4Q67L GTPase in complex with the central Rab binding domain of Rabenosyn-5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FYVE-finger-containing Rab5 effector protein rabenosyn-5, GTP-binding protein, ... | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-01 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1Z22
 
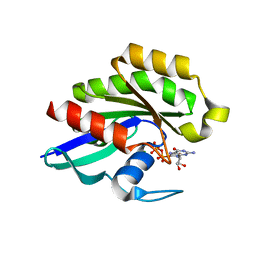 | | GDP-Bound Rab23 GTPase crystallized in C222(1) space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-23 | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-07 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
1Z0A
 
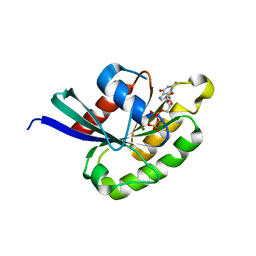 | | GDP-Bound Rab2A GTPase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-2A | | Authors: | Eathiraj, S, Pan, X, Ritacco, C, Lambright, D.G. | | Deposit date: | 2005-03-01 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis of family-wide Rab GTPase recognition by rabenosyn-5.
Nature, 436, 2005
|
|
