4LF2
 
 | | Hexameric Form II RuBisCO from Rhodopseudomonas palustris, activated and complexed with sulfate and magnesium | | Descriptor: | CARBONATE ION, MAGNESIUM ION, Ribulose bisphosphate carboxylase, ... | | Authors: | Chan, S, Satagopan, S, Sawaya, M.R, Eisenberg, D, Tabita, F.R, Perry, L.J. | | Deposit date: | 2013-06-26 | | Release date: | 2014-06-25 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure-function studies with the unique hexameric form II ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Rhodopseudomonas palustris.
J.Biol.Chem., 289, 2014
|
|
6HJM
 
 | |
5IRA
 
 | | Expanding Nature's Catalytic Repertoire -Directed Evolution of an Artificial Metalloenzyme for In Vivo Metathesis | | Descriptor: | Artificial Metathesase, [1-[4-[[5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]methyl]-2,6-dimethyl-phenyl]-3-(2,4,6-trimethylphenyl)-4,5-dihydroimidazol-1-ium-2-yl]-bis(chloranyl)ruthenium | | Authors: | Heinisch, T, Jeschek, M, Reuter, R, Trindler, C, Panke, S, Ward, T.R. | | Deposit date: | 2016-03-12 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Directed evolution of artificial metalloenzymes for in vivo metathesis.
Nature, 537, 2016
|
|
6CQG
 
 | |
5XAY
 
 | |
8E7F
 
 | | Crystal structure of the autotransporter Ssp from Serratia marcescens. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Extracellular serine protease, ... | | Authors: | Hor, L, Pilapitiya, A, Panjikar, S, Paxman, J.J, Heras, B. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a subtilisin-like autotransporter passenger domain reveals insights into its cytotoxic function.
Nat Commun, 14, 2023
|
|
5XAZ
 
 | |
6AC3
 
 | |
3GZJ
 
 | | Crystal Structure of Polyneuridine Aldehyde Esterase Complexed with 16-epi-Vellosimine | | Descriptor: | 16-epi-Vellosimine, Polyneuridine-aldehyde esterase | | Authors: | Yang, L, Hill, M, Wang, M, Panjikar, S, Stoeckigt, J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis and enzymatic mechanism of the biosynthesis of C9- from C10-monoterpenoid indole alkaloids
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5Y48
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4-dione at 1.70 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, URACIL | | Authors: | Singh, P.K, Pandey, S, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
5WV1
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with ribose sugar at 1.90 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Shokeen, A, Singh, P.K, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with ribose sugar at
1.90 A resolution.
To Be Published
|
|
3K3D
 
 | |
2HAK
 
 | | Catalytic and ubiqutin-associated domains of MARK1/PAR-1 | | Descriptor: | Serine/threonine-protein kinase MARK1 | | Authors: | Marx, A, Nugoor, C, Mueller, J, Panneerselvam, S, Mandelkow, E.-M, Mandelkow, E. | | Deposit date: | 2006-06-13 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variations in the catalytic and ubiquitin-associated domains of microtubule-associated protein/microtubule affinity regulating kinase (MARK) 1 and MARK2
J.Biol.Chem., 281, 2006
|
|
3KE6
 
 | |
1FBZ
 
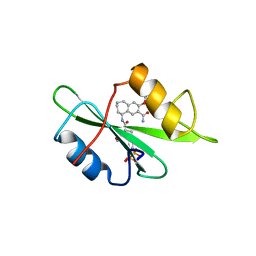 | | Structure-based design of a novel, osteoclast-selective, nonpeptide Src SH2 inhibitor with in vivo anti-resorptive activity | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK, {4-[2-ACETYLAMINO-2-(3-CARBAMOYL-2-CYCLOHEXYLMETHOXY-6,7,8,9-TETRAHYDRO-5H-BENZOCYCLOHEPTEN-5YLCARBAMOYL)-ETHYL]-2-PHOSPHONO-PHENYL}-PHOSPHONIC ACID | | Authors: | Shakespeare, W, Yang, M, Bohacek, R, Cerasoli, F, Stebbis, K, Sundaramoorthi, R, Vu, C, Pradeepan, S, Metcalf, C, Haraldson, C, Merry, T, Dalgarno, D, Narula, S, Hatada, M, Lu, X, Van Schravendijk, M.R, Adams, S, Violette, S, Smith, J, Guan, W, Bartlett, C, Herson, J, Iuliucci, J, Weigele, M, Sawyer, T. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of an osteoclast-selective, nonpeptide src homology 2 inhibitor with in vivo antiresorptive activity.
Proc.Natl.Acad.Sci.Usa, 97, 2000
|
|
2ZBB
 
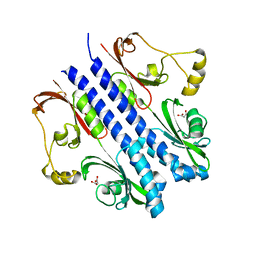 | | P43 crystal of DctBp | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, MALONIC ACID | | Authors: | Zhou, Y.F, Nan, B.Y, Liu, X, Nan, J, Liang, Y.H, Panjikar, S, Ma, Q.J, Wang, Y.P, Su, X.-D. | | Deposit date: | 2007-10-18 | | Release date: | 2008-11-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | crystal structures of sensory histidine kinase DctBp
to be published
|
|
2HLH
 
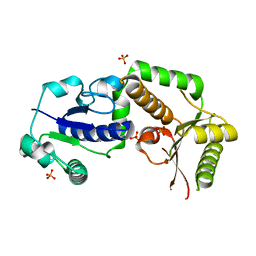 | | Crystal structure of fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | Nodulation fucosyltransferase, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
2HHC
 
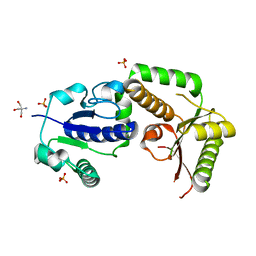 | | Crystal structure of fucosyltransferase NodZ from Bradyrhizobium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Stepkowski, T, Panjikar, S, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-06-28 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | High-resolution structure of NodZ fucosyltransferase involved in the biosynthesis of the nodulation factor.
Acta Biochim.Pol., 54, 2007
|
|
2B5S
 
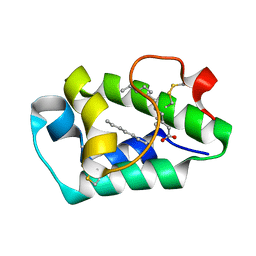 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, LAURIC ACID, Non-specific lipid transfer protein, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, R.J, Zanotti, G. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
2ALG
 
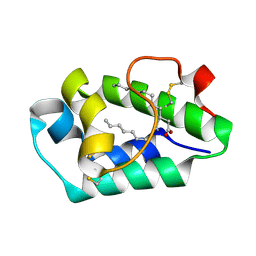 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, HEXAETHYLENE GLYCOL, LAURIC ACID, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, J, Zanotti, G. | | Deposit date: | 2005-08-05 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
2LHU
 
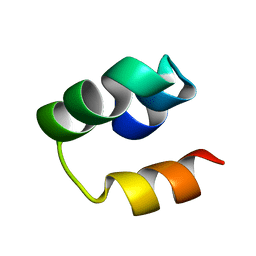 | | Structural Insight into the Unique Cardiac Myosin Binding Protein-C Motif: A Partially Folded Domain | | Descriptor: | Mybpc3 protein | | Authors: | Howarth, J.W, Rosevear, P.R, Ramisetti, S, Nolan, K, Sadayappan, S. | | Deposit date: | 2011-08-18 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Unique Cardiac Myosin-binding Protein-C Motif: A PARTIALLY FOLDED DOMAIN.
J.Biol.Chem., 287, 2012
|
|
2YKF
 
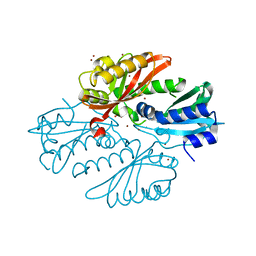 | | Sensor region of a sensor histidine kinase | | Descriptor: | BROMIDE ION, PROBABLE SENSOR HISTIDINE KINASE PDTAS, SULFATE ION | | Authors: | Preu, J, Panjikar, S, Morth, P, Jaiswal, R, Karunakar, P, Tucker, P.A. | | Deposit date: | 2011-05-26 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Sensor Region of the Ubiquitous Cytosolic Sensor Kinase, Pdtas, Contains Pas and Gaf Domain Sensing Modules.
J.Struct.Biol., 177, 2012
|
|
2Y9M
 
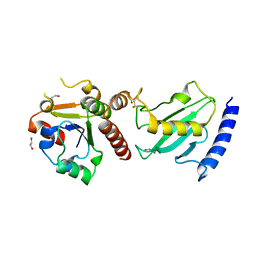 | | Pex4p-Pex22p structure | | Descriptor: | 1,2-ETHANEDIOL, PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-10-26 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into Ubiquitin-Conjugating Enzyme/ Co-Activator Interactions from the Structure of the Pex4P:Pex22P Complex.
Embo J., 31, 2011
|
|
2YFQ
 
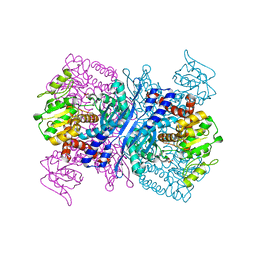 | | Crystal structure of Glutamate dehydrogenase from Peptoniphilus asaccharolyticus | | Descriptor: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE, SULFATE ION | | Authors: | Oliveira, T, Panjikar, S, Carrigan, J.B, Sharkey, M.A, Hamza, M, Engel, P.C, Khan, A.R. | | Deposit date: | 2011-04-07 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal Structure of Nad-Dependent Peptoniphilus Asaccharolyticus Glutamate Dehydrogenase Reveals Determinants of Cofactor Specificity.
J.Struct.Biol., 177, 2012
|
|
2Y9P
 
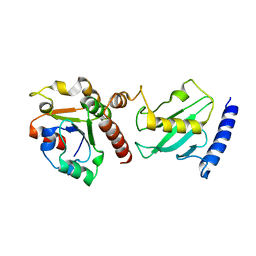 | | Pex4p-Pex22p mutant II structure | | Descriptor: | PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Pex4P-Pex22P Structure
To be Published
|
|
