8FSZ
 
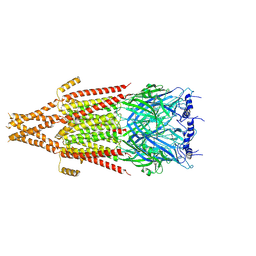 | | Full-length mouse 5-HT3A receptor in complex with ALB148471, open-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,5R)-1-azabicyclo[3.2.2]nonan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4EK7
 
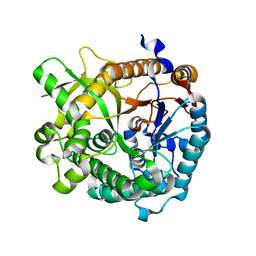 | | High speed X-ray analysis of plant enzymes at room temperature | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, beta-D-glucopyranose | | Authors: | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2012-04-09 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High speed X-ray analysis of plant enzymes at room temperature.
Phytochemistry, 2012
|
|
4EFQ
 
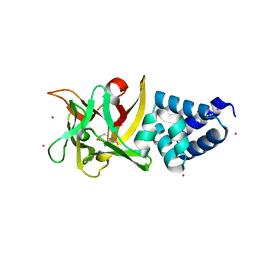 | | Bombyx mori lipoprotein 7 - platinum derivative at 1.94 A resolution | | Descriptor: | 30kDa protein, PLATINUM (II) ION, POTASSIUM ION, ... | | Authors: | Pietrzyk, A.J, Panjikar, S, Bujacz, A, Mueller-Dieckmann, J, Jaskolski, M, Bujacz, G. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | High-resolution structure of Bombyx mori lipoprotein 7: crystallographic determination of the identity of the protein and its potential role in detoxification.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EFP
 
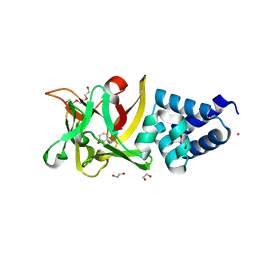 | | Bombyx mori lipoprotein 7 isolated from its natural source at 1.33 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 30kDa protein, CADMIUM ION, ... | | Authors: | Pietrzyk, A.J, Panjikar, S, Bujacz, A, Mueller-Dieckmann, J, Jaskolski, M, Bujacz, G. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-29 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-resolution structure of Bombyx mori lipoprotein 7: crystallographic determination of the identity of the protein and its potential role in detoxification.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6EBB
 
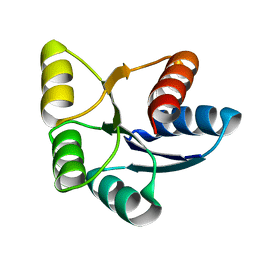 | |
5TRU
 
 | | Structure of the first-in-class checkpoint inhibitor Ipilimumab bound to human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Ipilimumab Fab heavy chain, Ipilimumab Fab light chain | | Authors: | Ramagopal, U.A, Liu, W, Garrett-Thomson, S.C, Yan, Q, Srinivasan, M, Wong, S.C, Bell, A, Mankikar, S, Rangan, V.S, Deshpande, S, Bonanno, J.B, Korman, A.J, Almo, S.C. | | Deposit date: | 2016-10-27 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2VAQ
 
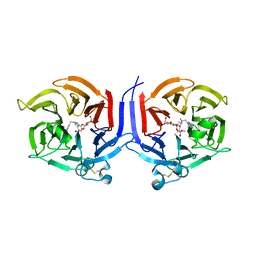 | | STRUCTURE OF STRICTOSIDINE SYNTHASE IN COMPLEX WITH INHIBITOR | | Descriptor: | (2S,3R,4S)-methyl 4-(2-(2-(1H-indol-3-yl)ethylamino)ethyl)-2-((2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yloxy)-3-vinyl-3,4-dihydro-2H-pyran-5-carboxylate, STRICTOSIDINE SYNTHASE | | Authors: | Maresh, J, Giddings, L.A, Friedrich, A, Loris, E.A, Panjikar, S, Trout, B.L, Stoeckigt, J, Peters, B, O'Connor, S.E. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Strictosidine synthase: mechanism of a Pictet-Spengler catalyzing enzyme.
J. Am. Chem. Soc., 130, 2008
|
|
6FH8
 
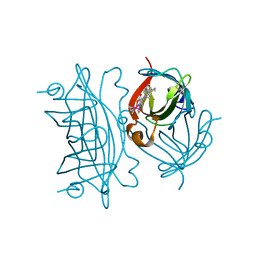 | | E. coli surface display of streptavidin for directed evolution of an allylic deallocase | | Descriptor: | Streptavidin, biotinylated ruthenium cyclopentadienide | | Authors: | Heinisch, T, Schwizer, F, Garabedian, B, Csibra, E, Jeschek, M, Pinheiro Bernhardes, V, Marliere, P, Panke, S, Ward, T.R. | | Deposit date: | 2018-01-12 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | E. colisurface display of streptavidin for directed evolution of an allylic deallylase.
Chem Sci, 9, 2018
|
|
1PZM
 
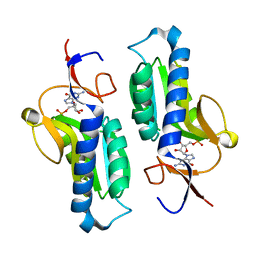 | | Crystal structure of HGPRT-ase from Leishmania tarentolae in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Monzani, P.S, Trapani, S, Oliva, G, Thiemann, O.H. | | Deposit date: | 2003-07-11 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Leishmania tarentolae hypoxanthine-guanine phosphoribosyltransferase.
Bmc Struct.Biol., 7, 2007
|
|
2WFM
 
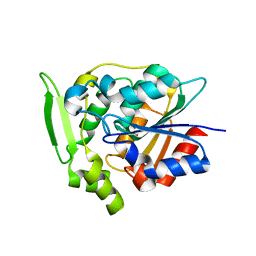 | | Crystal structure of polyneuridine aldehyde esterase mutant (H244A) | | Descriptor: | POLYNEURIDINE ALDEHYDE ESTERASE | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1NWA
 
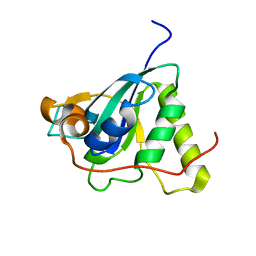 | | Structure of Mycobacterium tuberculosis Methionine Sulfoxide Reductase A in Complex with Protein-bound Methionine | | Descriptor: | Peptide methionine sulfoxide reductase msrA | | Authors: | Taylor, A.B, Benglis Jr, D.M, Dhandayuthapani, S, Hart, P.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Mycobacterium tuberculosis Methionine Sulfoxide Reductase A in Complex with Protein-bound Methionine
J.Bacteriol., 185, 2003
|
|
2W00
 
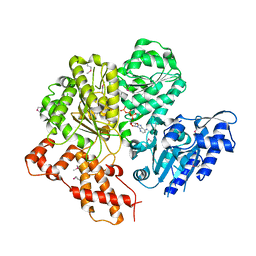 | | Crystal structure of the HsdR subunit of the EcoR124I restriction enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HSDR, MAGNESIUM ION | | Authors: | Lapkouski, M, Panjikar, S, Kuta Smatanova, I, Ettrich, R, Csefalvay, E. | | Deposit date: | 2008-08-08 | | Release date: | 2008-12-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Motor Subunit of Type I Restriction-Modification Complex Ecor124I.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1MVQ
 
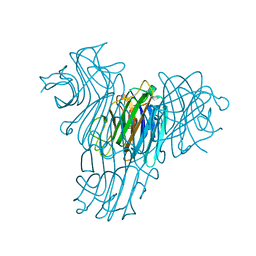 | | Cratylia mollis lectin (isoform 1) in complex with methyl-alpha-D-mannose | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, lectin, ... | | Authors: | de Souza, G.A, Oliveira, P.S, Trapani, S, Correia, M.T, Oliva, G, Coelho, L.C, Greene, L.J. | | Deposit date: | 2002-09-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Amino acid sequence and tertiary structure of Cratylia mollis seed lectin
Glycobiology, 13, 2003
|
|
4HGH
 
 | | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset I) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
2WFL
 
 | | Crystal structure of polyneuridine aldehyde esterase | | Descriptor: | POLYNEURIDINE-ALDEHYDE ESTERASE, SULFATE ION | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4HGG
 
 | | Crystal structure of P450 BM3 5F5R heme domain variant complexed with styrene | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGJ
 
 | | Crystal structure of P450 BM3 5F5 heme domain variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGF
 
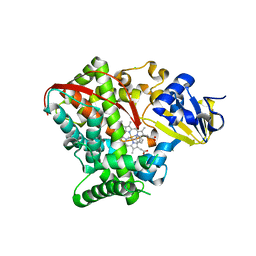 | | Crystal structure of P450 BM3 5F5K heme domain variant complexed with styrene | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGI
 
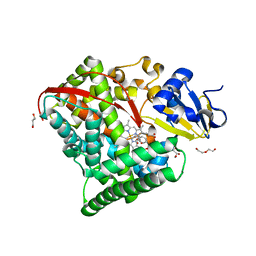 | | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset II) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
1O1H
 
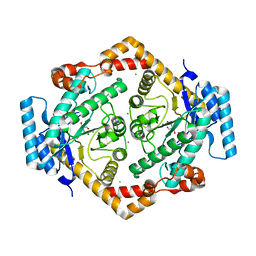 | | STRUCTURE OF GLUCOSE ISOMERASE DERIVATIZED WITH KR. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Nowak, E, Panjikar, S, Tucker, P.A. | | Deposit date: | 2002-11-07 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Glucose Isomerase Derivatized with Kr.
To be Published
|
|
5FA6
 
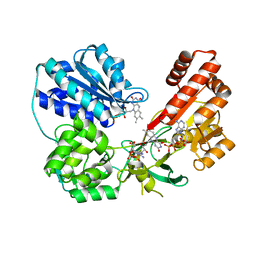 | | wild type human CYPOR | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Marohnic, C, Panda, S, Masters, B.S, Kim, J.J.K. | | Deposit date: | 2015-12-11 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Instability of the Human Cytochrome P450 Reductase A287P Variant Is the Major Contributor to Its Antley-Bixler Syndrome-like Phenotype.
J.Biol.Chem., 291, 2016
|
|
4GUV
 
 | | TetX derivatized with Xenon | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TetX2 protein, ... | | Authors: | Volkers, G, Palm, G.J, Panjikar, S, Hinrichs, W. | | Deposit date: | 2012-08-29 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Putative dioxygen-binding sites and recognition of tigecycline and minocycline in the tetracycline-degrading monooxygenase TetX.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2YFH
 
 | | Structure of a Chimeric Glutamate Dehydrogenase | | Descriptor: | GLUTAMATE DEHYDROGENASE, NAD-SPECIFIC GLUTAMATE DEHYDROGENASE | | Authors: | Oliveira, T, Panjikar, S, Sharkey, M.A, Carrigan, J.B, Hamza, M, Engel, P.C, Khan, A.R. | | Deposit date: | 2011-04-05 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of a Chimaeric Bacterial Glutamate Dehydrogenase.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2XTS
 
 | | Crystal Structure of the Sulfane Dehydrogenase SoxCD from Paracoccus pantotrophus | | Descriptor: | CALCIUM ION, COBALT (II) ION, CYTOCHROME, ... | | Authors: | Zander, U, Faust, A, Klink, B.U, de Sanctis, D, Panjikar, S, Quentmeier, A, Bardischewski, F, Friedrich, C.G, Scheidig, A.J. | | Deposit date: | 2010-10-12 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural Basis for the Oxidation of Protein-Bound Sulfur by the Sulfur Cycle Molybdohemo-Enzyme Sulfane Dehydrogenase Soxcd.
J.Biol.Chem., 286, 2011
|
|
2LHU
 
 | | Structural Insight into the Unique Cardiac Myosin Binding Protein-C Motif: A Partially Folded Domain | | Descriptor: | Mybpc3 protein | | Authors: | Howarth, J.W, Rosevear, P.R, Ramisetti, S, Nolan, K, Sadayappan, S. | | Deposit date: | 2011-08-18 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Unique Cardiac Myosin-binding Protein-C Motif: A PARTIALLY FOLDED DOMAIN.
J.Biol.Chem., 287, 2012
|
|
