5MYY
 
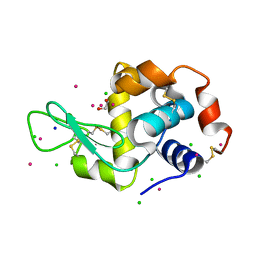 | | Hen Egg-White Lysozyme (HEWL) cocrystallized in the presence of Cadmium sulphate | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Panneerselvam, S, Burkhardt, A, Meents, A. | | Deposit date: | 2017-01-30 | | Release date: | 2017-07-12 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rapid cadmium SAD phasing at the standard wavelength (1 angstrom ).
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2WZJ
 
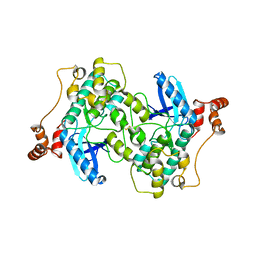 | | Catalytic and UBA domain of kinase MARK2/(Par-1) K82R, T208E double mutant | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE MARK2 | | Authors: | Panneerselvam, S, Marx, A, Mandelkow, E.-M, Mandelkow, E. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | Structure and Function of Polarity-Inducing Kinase Family Mark/Par-1 within the Branch of Ampk/Snf1-Related Kinases.
Faseb J., 24, 2010
|
|
5JW8
 
 | |
7FHZ
 
 | |
7FI1
 
 | |
7FI2
 
 | |
7FI0
 
 | | Crystal structure of Multi-functional Polysaccharide lyase Smlt1473 (WT) from Stenotrophomonas maltophilia (strain K279a) in ManA bound form at pH-5.0 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Polysaccharide lyase, SULFATE ION, ... | | Authors: | Pandey, S, Berger, B.W, Acharya, R. | | Deposit date: | 2021-07-30 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural insights into the mechanism of pH-selective substrate specificity of the polysaccharide lyase Smlt1473.
J.Biol.Chem., 297, 2021
|
|
7FHY
 
 | |
7FHX
 
 | |
7FHU
 
 | |
7FHV
 
 | |
7FHW
 
 | |
1OZG
 
 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate | | Descriptor: | 2-HYDROXYETHYL DIHYDROTHIACHROME DIPHOSPHATE, Acetolactate synthase, catabolic, ... | | Authors: | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | Deposit date: | 2003-04-09 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
7ZC9
 
 | | Human Pikachurin/EGFLAM C-terminal Laminin-G domain (LG3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pikachurin, SULFATE ION | | Authors: | Pantalone, S, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
7ZCB
 
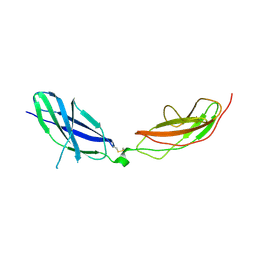 | | Human Pikachurin/EGFLAM N-terminal Fibronectin-III (1-2) domains | | Descriptor: | CHLORIDE ION, Pikachurin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pantalone, S, Savino, S, Viti, L.V, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
1OZF
 
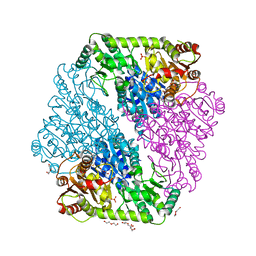 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactors | | Descriptor: | Acetolactate synthase, catabolic, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | Deposit date: | 2003-04-09 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
1OZH
 
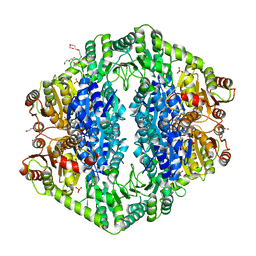 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate. | | Descriptor: | 2-HYDROXYETHYL DIHYDROTHIACHROME DIPHOSPHATE, Acetolactate synthase, catabolic, ... | | Authors: | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | Deposit date: | 2003-04-09 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
1ZMV
 
 | |
2R0I
 
 | |
1Y8G
 
 | |
2BD0
 
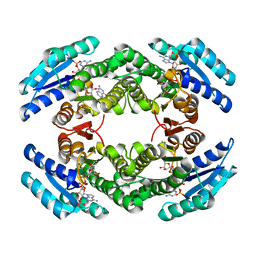 | | Chlorobium tepidum Sepiapterin Reductase complexed with NADP and Sepiapterin | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, sepiapterin reductase | | Authors: | Supangat, S, Seo, K.H, Choi, Y.K, Park, Y.S, Lee, K.H. | | Deposit date: | 2005-10-19 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Chlorobium tepidum sepiapterin reductase complex reveals the novel substrate binding mode for stereospecific production of L-threo-tetrahydrobiopterin
J.Biol.Chem., 281, 2006
|
|
1ZMW
 
 | |
1ZMU
 
 | |
7ZF2
 
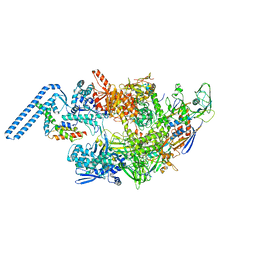 | | Protomeric substructure from an octameric assembly of M. tuberculosis RNA polymerase in complex with sigma-b initiation factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Trapani, S, Bron, P, Lai Kee Him, J, Brodolin, K, Morichaud, Z, Vishwakarma, R. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization.
Nat Commun, 14, 2023
|
|
1L1G
 
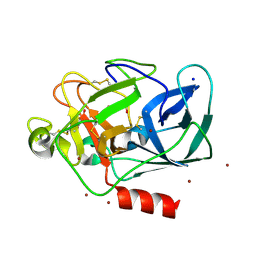 | | The Structure of Porcine Pancreatic Elastase Complexed with Xenon and Bromide, Cryoprotected with Glycerol | | Descriptor: | BROMIDE ION, ELASTASE 1, GLYCEROL, ... | | Authors: | Panjikar, S, Tucker, P.A. | | Deposit date: | 2002-02-16 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Xenon derivatization of halide-soaked protein crystals.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
