7RQP
 
 | |
7RQR
 
 | |
5UTY
 
 | | Crystal Structure of a Stabilized DS-SOSIP.mut4 BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, N300M, N302M, T320L in Complex with Human Antibodies PGT122 and 35O22 at 4.1 Angstrom | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | 著者 | Xu, K, Chuang, G.-Y, Pancera, M, Kwong, P.D. | | 登録日 | 2017-02-15 | | 公開日 | 2017-03-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.412 Å) | | 主引用文献 | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
1GPZ
 
 | | THE CRYSTAL STRUCTURE OF THE ZYMOGEN CATALYTIC DOMAIN OF COMPLEMENT PROTEASE C1R | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C1R COMPONENT, ... | | 著者 | Budayova-Spano, M, Fontecilla-Camps, J.C, Gaboriaud, C. | | 登録日 | 2001-11-15 | | 公開日 | 2002-07-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Crystal Structure of the Zymogen Catalytic Domain of Complement Protease C1R Reveals that a Disruptive Mechanical Stress is Required to Trigger Activation of the C1 Complex.
Embo J., 21, 2002
|
|
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | 著者 | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | 登録日 | 2009-05-18 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IDX
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C222 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab b13 heavy chain, ... | | 著者 | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | 登録日 | 2009-07-22 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IDY
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C2221 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab b13 heavy chain, Fab b13 light chain, ... | | 著者 | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | 登録日 | 2009-07-22 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
4N9M
 
 | | Joint neutron/x-ray structure of urate oxidase in complex with 8-hydroxyxanthine | | 分子名称: | 8-hydroxy-3,9-dihydro-1H-purine-2,6-dione, CHLORIDE ION, SODIUM ION, ... | | 著者 | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | 登録日 | 2013-10-21 | | 公開日 | 2014-02-05 | | 最終更新日 | 2018-06-13 | | 実験手法 | NEUTRON DIFFRACTION (2.023 Å), X-RAY DIFFRACTION | | 主引用文献 | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
4N9V
 
 | | High resolution x-ray structure of urate oxidase in complex with 8-azaxanthine | | 分子名称: | 8-AZAXANTHINE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | 登録日 | 2013-10-21 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
3FRH
 
 | | Structure of the 16S rRNA methylase RmtB, P21 | | 分子名称: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | 登録日 | 2009-01-08 | | 公開日 | 2009-08-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
3FRI
 
 | | Structure of the 16S rRNA methylase RmtB, I222 | | 分子名称: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | 登録日 | 2009-01-08 | | 公開日 | 2009-08-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
3FZG
 
 | | Structure of the 16S rRNA methylase ArmA | | 分子名称: | 16S rRNA methylase, S-ADENOSYLMETHIONINE | | 著者 | Schmitt, E, Galimand, M, Panvert, M, Courvalin, P, Mechulam, Y. | | 登録日 | 2009-01-26 | | 公開日 | 2009-08-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
7RAQ
 
 | |
7RDW
 
 | |
4N9S
 
 | | High resolution X-RAY STRUCTURE OF URATE OXIDASE IN COMPLEX WITH 8-HYDROXYXANTHINE | | 分子名称: | 8-hydroxy-3,9-dihydro-1H-purine-2,6-dione, CHLORIDE ION, GLYCEROL, ... | | 著者 | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | 登録日 | 2013-10-21 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
4N3M
 
 | | Joint neutron/X-ray structure of urate oxidase in complex with 8-azaxanthine | | 分子名称: | 8-AZAXANTHINE, CHLORIDE ION, SODIUM ION, ... | | 著者 | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | 登録日 | 2013-10-07 | | 公開日 | 2014-02-05 | | 最終更新日 | 2018-06-13 | | 実験手法 | NEUTRON DIFFRACTION (1.919 Å), X-RAY DIFFRACTION | | 主引用文献 | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
1M3D
 
 | | Structure of Type IV Collagen NC1 Domains | | 分子名称: | BROMIDE ION, GLYCEROL, LUTETIUM (III) ION, ... | | 著者 | Sundaramoorthy, M, Meiyappan, M, Todd, P, Hudson, B.G. | | 登録日 | 2002-06-27 | | 公開日 | 2003-01-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of NC1 Domains. Structural Basis for Type IV Collagen Assembly in Basement Membranes
J.Biol.Chem., 277, 2002
|
|
9DR0
 
 | |
9DQC
 
 | |
8V96
 
 | | GII.14 M7 norovirus protruding domain | | 分子名称: | 1,2-ETHANEDIOL, Capsid protein VP1 | | 著者 | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | 登録日 | 2023-12-07 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
2WOO
 
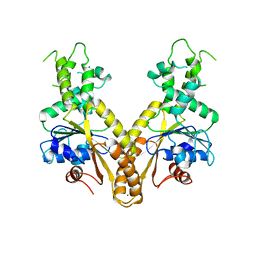 | | Nucleotide-free form of S. pombe Get3 | | 分子名称: | ATPASE GET3, ZINC ION | | 著者 | Mateja, A, Szlachcic, A, Downing, M.E, Dobosz, M, Mariappan, M, Hegde, R.S, Keenan, R.J. | | 登録日 | 2009-07-27 | | 公開日 | 2009-08-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.006 Å) | | 主引用文献 | The Structural Basis of Tail-Anchored Membrane Protein Recognition by Get3.
Nature, 461, 2009
|
|
2WOJ
 
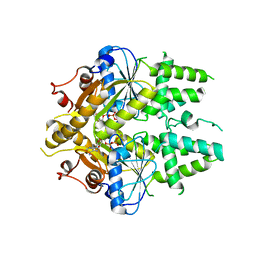 | | ADP-AlF4 complex of S. cerevisiae GET3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATPASE GET3, MAGNESIUM ION, ... | | 著者 | Mateja, A, Szlachcic, A, Downing, M.E, Dobosz, M, Mariappan, M, Hegde, R.S, Keenan, R.J. | | 登録日 | 2009-07-26 | | 公開日 | 2009-08-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.994 Å) | | 主引用文献 | The Structural Basis of Tail-Anchored Membrane Protein Recognition by Get3.
Nature, 461, 2009
|
|
8V98
 
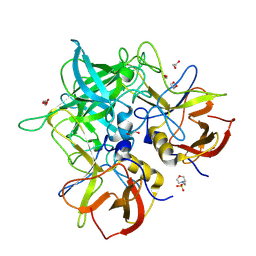 | | GII.24 Loreto 1972 norovirus protruding domain | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Capsid protein VP1 | | 著者 | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | 登録日 | 2023-12-07 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
8V9A
 
 | | GII.NA1 Loreto 1257 norovirus protruding domain | | 分子名称: | 1,2-ETHANEDIOL, Capsid protein VP1 | | 著者 | Kher, G, Kim, I, Pancera, M, Hansman, G. | | 登録日 | 2023-12-07 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
8V97
 
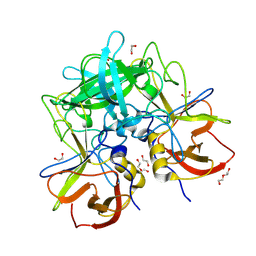 | | GII.17 CS-E1 norovirus protruding domain | | 分子名称: | 1,2-ETHANEDIOL, Capsid protein VP1, DI(HYDROXYETHYL)ETHER | | 著者 | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | 登録日 | 2023-12-07 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
