3KX0
 
 | | Crystal Structure of the PAS domain of Rv1364c | | Descriptor: | ISOPROPYL ALCOHOL, Uncharacterized protein Rv1364c/MT1410 | | Authors: | Jaiswal, R.K, Gopal, B. | | Deposit date: | 2009-12-02 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a PAS sensor domain in the Mycobacterium tuberculosis transcription regulator Rv1364c
Biochem.Biophys.Res.Commun., 398, 2010
|
|
3KHZ
 
 | |
3KHX
 
 | |
3KI9
 
 | |
4BXI
 
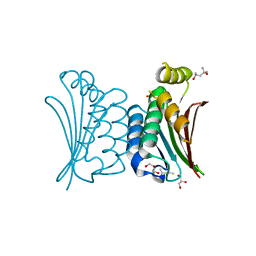 | | Crystal structure of ATP binding domain of AgrC from Staphylococcus aureus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACCESSORY GENE REGULATOR PROTEIN C, ACETATE ION, ... | | Authors: | Srivastava, S.K, Rajasree, K, Gopal, B. | | Deposit date: | 2013-07-12 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Influence of the Agrc-Agra Complex in the Response Time of Staphylococcus Aureus Quorum Sensing
J.Bacteriol., 196, 2014
|
|
3H7Y
 
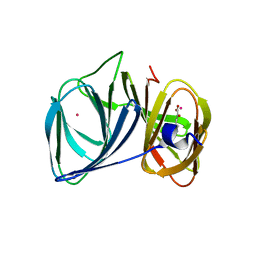 | | Crystal structure of BacB, an enzyme involved in Bacilysin synthesis, in tetragonal form | | Descriptor: | 3-PHENYLPYRUVIC ACID, Bacilysin biosynthesis protein bacB, COBALT (II) ION, ... | | Authors: | Rajavel, M, Gopal, B. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Role of Bacillus subtilis BacB in the synthesis of bacilysin
J.Biol.Chem., 284, 2009
|
|
3H7J
 
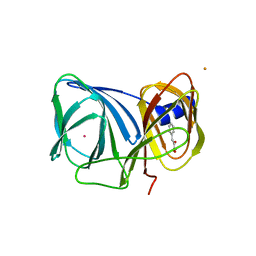 | | Crystal structure of BacB, an enzyme involved in Bacilysin synthesis, in monoclinic form | | Descriptor: | 3-PHENYLPYRUVIC ACID, Bacilysin biosynthesis protein bacB, COBALT (II) ION, ... | | Authors: | Rajavel, M, Gopal, B. | | Deposit date: | 2009-04-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Role of Bacillus subtilis BacB in the synthesis of bacilysin
J.Biol.Chem., 284, 2009
|
|
2PI7
 
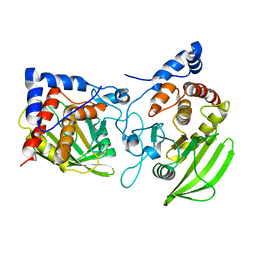 | |
4NQW
 
 | |
3S3K
 
 | |
3S3E
 
 | |
3VFZ
 
 | |
4PG4
 
 | |
4PG8
 
 | |
3U4C
 
 | |
3VEP
 
 | | Crystal structure of SigD4 in complex with its negative regulator RsdA | | Descriptor: | Probable RNA polymerase sigma-D factor, SULFATE ION, Uncharacterized protein Rv3413c/MT3522 | | Authors: | Jaiswal, R.K, Gopal, B. | | Deposit date: | 2012-01-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis RsdA provides a conformational rationale for selective regulation of sigma-factor activity by proteolysis
Nucleic Acids Res., 41, 2013
|
|
4PG6
 
 | |
3U4D
 
 | |
4PG5
 
 | |
4PG7
 
 | |
3W8W
 
 | | The crystal structure of EncM | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
3U49
 
 | |
3W8Z
 
 | | The complex structure of EncM with hydroxytetraketide | | Descriptor: | (7S)-7-hydroxy-1-phenyloctane-1,3,5-trione, FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
3W8X
 
 | | The complex structure of EncM with trifluorotriketide | | Descriptor: | 6,6,6-trifluoro-1-phenylhexane-1,3,5-trione, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
3Q89
 
 | |
