8I6A
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Orotic acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, OROTIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I6C
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 6-Formyl-uracil, Form III | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I6D
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Hydroxy-2,4(1H,3H)-pyrimidinedione, Form VI | | Descriptor: | 1,2-ETHANEDIOL, 5-oxidanyl-1~{H}-pyrimidine-2,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I64
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid, Form II | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I68
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Uric acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, URIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
5ITW
 
 | | Crystal structure of Bacillus subtilis BacC Dihydroanticapsin 7-dehydrogenase | | Descriptor: | Dihydroanticapsin 7-dehydrogenase, SULFATE ION | | Authors: | Perinbam, K, Balaram, H, Row, T.N.G, Gopal, B. | | Deposit date: | 2016-03-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Probing the influence of non-covalent contact networks identified by charge density analysis on the oxidoreductase BacC.
Protein Eng. Des. Sel., 30, 2017
|
|
5ITV
 
 | | Crystal structure of Bacillus subtilis BacC Dihydroanticapsin 7-dehydrogenase in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dihydroanticapsin 7-dehydrogenase | | Authors: | Perinbam, K, Balaram, H, Row, T.N.G, Gopal, B. | | Deposit date: | 2016-03-17 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Probing the influence of non-covalent contact networks identified by charge density analysis on the oxidoreductase BacC.
Protein Eng. Des. Sel., 30, 2017
|
|
5JFE
 
 | | Flavin-dependent thymidylate synthase with H2-dUMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-uridylic acid, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sapra, A, Stuckey, J, Palfey, B. | | Deposit date: | 2016-04-19 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Evaluating H2-dUMP as an Intermediate in the oxidation of Flavin-dependent Thymidylate Synthase
To Be Published
|
|
5J5D
 
 | |
7VZ0
 
 | | Structure of the M. tuberculosis HtrA S407A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7VYZ
 
 | | Structure of the M. tuberculosis HtrA S367A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
6IEO
 
 | |
3H7J
 
 | | Crystal structure of BacB, an enzyme involved in Bacilysin synthesis, in monoclinic form | | Descriptor: | 3-PHENYLPYRUVIC ACID, Bacilysin biosynthesis protein bacB, COBALT (II) ION, ... | | Authors: | Rajavel, M, Gopal, B. | | Deposit date: | 2009-04-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Role of Bacillus subtilis BacB in the synthesis of bacilysin
J.Biol.Chem., 284, 2009
|
|
3H7Y
 
 | | Crystal structure of BacB, an enzyme involved in Bacilysin synthesis, in tetragonal form | | Descriptor: | 3-PHENYLPYRUVIC ACID, Bacilysin biosynthesis protein bacB, COBALT (II) ION, ... | | Authors: | Rajavel, M, Gopal, B. | | Deposit date: | 2009-04-28 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Role of Bacillus subtilis BacB in the synthesis of bacilysin
J.Biol.Chem., 284, 2009
|
|
7RDN
 
 | | Crystal structure of S. cerevisiae pre-mRNA leakage protein 39 (Pml39) | | Descriptor: | Pre-mRNA leakage protein 39, ZINC ION | | Authors: | Hashimoto, H, Ramirez, D.H, Pawlak, N, Blobel, G, Palancade, B, Debler, E.W. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of the pre-mRNA leakage 39-kDa protein reveals a single domain of integrated zf-C3HC and Rsm1 modules.
Sci Rep, 12, 2022
|
|
4EWT
 
 | | The crystal structure of a putative aminohydrolase from methicillin resistant Staphylococcus aureus | | Descriptor: | 1-DEOXY-1-THIO-HEPTAETHYLENE GLYCOL, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Girish, T.S, Vivek, B, Colaco, M, Misquith, S, Gopal, B. | | Deposit date: | 2012-04-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an amidohydrolase, SACOL0085, from methicillin-resistant Staphylococcus aureus COL
Acta Crystallogr.,Sect.F, 69, 2013
|
|
8QLP
 
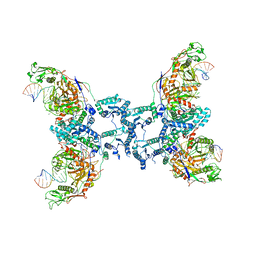 | | CryoEM structure of the RNA/DNA bound SPARTA (BabAgo/TIR-APAZ) tetrameric complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*C)-3'), MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3'), ... | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
8QLO
 
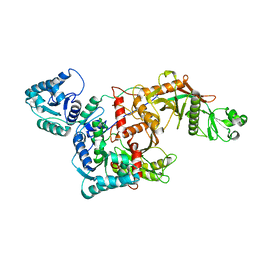 | | CryoEM structure of the apo SPARTA (BabAgo/TIR-APAZ) complex | | Descriptor: | Short prokaryotic Argonaute, Toll/interleukin-1 receptor domain-containing protein | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
3KX0
 
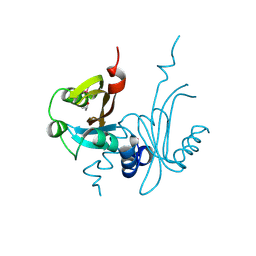 | | Crystal Structure of the PAS domain of Rv1364c | | Descriptor: | ISOPROPYL ALCOHOL, Uncharacterized protein Rv1364c/MT1410 | | Authors: | Jaiswal, R.K, Gopal, B. | | Deposit date: | 2009-12-02 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a PAS sensor domain in the Mycobacterium tuberculosis transcription regulator Rv1364c
Biochem.Biophys.Res.Commun., 398, 2010
|
|
3KHZ
 
 | |
3KI9
 
 | |
3KHX
 
 | |
2O7G
 
 | |
4BXI
 
 | | Crystal structure of ATP binding domain of AgrC from Staphylococcus aureus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACCESSORY GENE REGULATOR PROTEIN C, ACETATE ION, ... | | Authors: | Srivastava, S.K, Rajasree, K, Gopal, B. | | Deposit date: | 2013-07-12 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Influence of the Agrc-Agra Complex in the Response Time of Staphylococcus Aureus Quorum Sensing
J.Bacteriol., 196, 2014
|
|
2PI7
 
 | |
