1M8K
 
 | |
1LVW
 
 | | Crystal structure of glucose-1-phosphate thymidylyltransferase, RmlA, complex with dTDP | | 分子名称: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Dong, A, Christendat, D, Pai, E.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2002-05-29 | | 公開日 | 2003-07-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of glucose-1-phosphate thymidylyltransferase, RmlA, complex with dTDP
To be Published
|
|
1M8F
 
 | |
1M8J
 
 | |
3R3U
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - WT/apo | | 分子名称: | CHLORIDE ION, Fluoroacetate dehalogenase, NICKEL (II) ION | | 著者 | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | 登録日 | 2011-03-16 | | 公開日 | 2011-05-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3R41
 
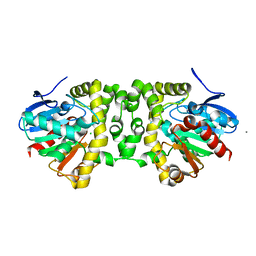 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/apo | | 分子名称: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase | | 著者 | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | 登録日 | 2011-03-17 | | 公開日 | 2011-05-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
1M8G
 
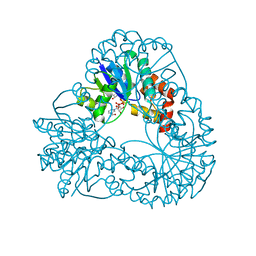 | |
3R3W
 
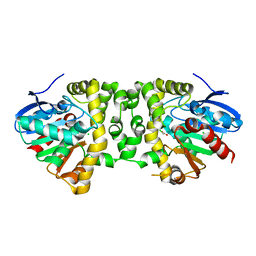 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Chloroacetate | | 分子名称: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase, ... | | 著者 | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | 登録日 | 2011-03-16 | | 公開日 | 2011-05-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3R3V
 
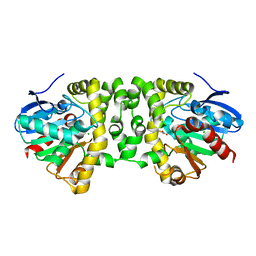 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Fluoroacetate | | 分子名称: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase, ... | | 著者 | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | 登録日 | 2011-03-16 | | 公開日 | 2011-05-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3RFN
 
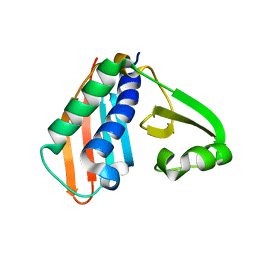 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | 分子名称: | BB_1wnu_001, ZINC ION | | 著者 | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | 登録日 | 2011-04-06 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RHU
 
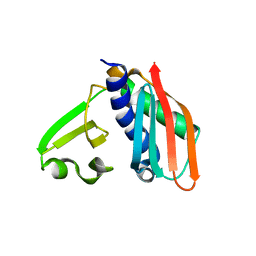 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | 分子名称: | SC_1wnu | | 著者 | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | 登録日 | 2011-04-12 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3R3Z
 
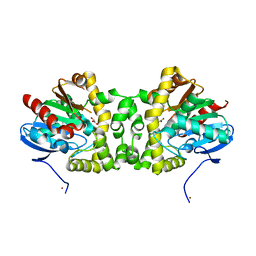 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - WT/Glycolate | | 分子名称: | Fluoroacetate dehalogenase, GLYCOLIC ACID, NICKEL (II) ION | | 著者 | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | 登録日 | 2011-03-16 | | 公開日 | 2011-05-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3R3Y
 
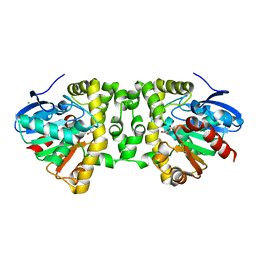 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate | | 分子名称: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase | | 著者 | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | 登録日 | 2011-03-16 | | 公開日 | 2011-05-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3RI0
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | 分子名称: | BB_2cx5_001, GLYCEROL, SULFATE ION | | 著者 | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | 登録日 | 2011-04-12 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3R40
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/apo | | 分子名称: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase | | 著者 | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | 登録日 | 2011-03-16 | | 公開日 | 2011-05-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3R3X
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Bromoacetate | | 分子名称: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase, ... | | 著者 | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | 登録日 | 2011-03-16 | | 公開日 | 2011-05-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
1N5X
 
 | | Xanthine Dehydrogenase from Bovine Milk with Inhibitor TEI-6720 Bound | | 分子名称: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Okamoto, K, Eger, B.T, Nishino, T, Kondo, S, Pai, E.F, Nishino, T. | | 登録日 | 2002-11-07 | | 公開日 | 2003-03-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | An Extremely Potent Inhibitor of Xanthine Oxidoreductase: Crystal Structure of the Enzyme-Inhibitor Complex and Mechanism of Inhibition
J.BIOL.CHEM., 278, 2003
|
|
3RIJ
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | 分子名称: | GLYCEROL, SC_2cx5 | | 著者 | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | 登録日 | 2011-04-13 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RZ4
 
 | |
3SGU
 
 | |
3S9Y
 
 | | Crystal Structure of P. falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP in space group P21, produced from 5-fluoro-6-azido-UMP | | 分子名称: | 6-amino-5-fluorouridine 5'-(dihydrogen phosphate), DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | 著者 | Liu, Y, Kotra, L.P, Pai, E.F. | | 登録日 | 2011-06-02 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
3SSJ
 
 | |
3SZE
 
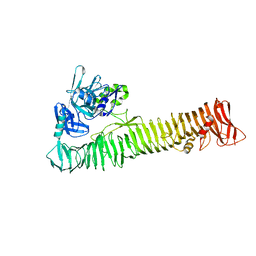 | | Crystal structure of the passenger domain of the E. coli autotransporter EspP | | 分子名称: | Serine protease espP | | 著者 | Khan, S, Mian, H.S, Sandercock, L.E, Battaile, K.P, Lam, R, Chirgadze, N.Y, Pai, E.F. | | 登録日 | 2011-07-18 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Passenger Domain of the Escherichia coli Autotransporter EspP.
J.Mol.Biol., 413, 2011
|
|
3SW6
 
 | |
3THQ
 
 | |
