4NEW
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma cruzi in complex with inhibitor EP127 (5-{5-[1-(PYRROLIDIN-1-YL)CYCLOHEXYL]-1,3-THIAZOL-2-YL}-1H-INDOLE) | | Descriptor: | 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, Trypanothione reductase, ... | | Authors: | Persch, E, Bryson, S, Pai, E.F, Krauth-Siegel, R.L, Diederich, F. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding to large enzyme pockets: small-molecule inhibitors of trypanothione reductase.
Chemmedchem, 9, 2014
|
|
3UMC
 
 | | Crystal Structure of the L-2-Haloacid Dehalogenase PA0810 | | Descriptor: | CHLORIDE ION, SODIUM ION, haloacid dehalogenase | | Authors: | Petit, P, Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-13 | | Release date: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
3UMB
 
 | | Crystal Structure of the L-2-Haloacid Dehalogenase RSc1362 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Petit, P, Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-12 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
3UMG
 
 | | Crystal Structure of the Defluorinating L-2-Haloacid Dehalogenase Rha0230 | | Descriptor: | CHLORIDE ION, Haloacid dehalogenase | | Authors: | Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-13 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
4PEO
 
 | | Crystal structure of a hypothetical protein from Staphylococcus aureus. | | Descriptor: | Hypothetical protein | | Authors: | McGrath, T.E, Kisselman, G, Romanov, V, Wu-Brown, J, Soloveychik, M, Chan, T.S.Y, Gordon, R.D, Thambipillai, D, Dharamsi, A, Mansoury, K, Battaile, K.P, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of a hypothetical protein from Staphylococcus aureus.
To Be Published
|
|
2ZZ7
 
 | |
2ZZ5
 
 | |
2ZZ3
 
 | |
2ZZ4
 
 | |
2ZZ2
 
 | |
2ZZ1
 
 | |
2ZZ6
 
 | |
3AMZ
 
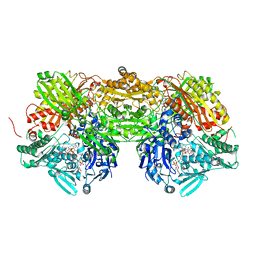 | | Bovine Xanthine Oxidoreductase urate bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Okamoto, K, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2010-08-27 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Urate Bound Form of Xanthine Oxidoreductase: Substrate Orientation and Structure of the Key Reaction Intermediate
J.Am.Chem.Soc., 132, 2010
|
|
3AN1
 
 | | Crystal structure of rat D428A mutant, urate bound form | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Kawaguchi, Y, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2010-08-27 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structures of Urate Bound Form of Xanthine Oxidoreductase: Substrate Orientation and Structure of the Key Reaction Intermediate
J.Am.Chem.Soc., 132, 2010
|
|
2ESH
 
 | | Crystal Structure of Conserved Protein of Unknown Function TM0937- a Potential Transcriptional Factor | | Descriptor: | CALCIUM ION, conserved hypothetical protein TM0937 | | Authors: | Liu, Y, Bochkareva, E, Zheng, H, Xu, X, Nocek, B, Lunin, V, Edward, A, Pai, E.F, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein TM0937
To be Published
|
|
3BGJ
 
 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 6-iodo-UMP | | Descriptor: | GLYCEROL, URIDINE-5'-MONOPHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Bello, A.M, Devalla, S, Kotra, L.P, Pai, E.F. | | Deposit date: | 2007-11-26 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 6-iodo-UMP
To be Published
|
|
421P
 
 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Krengel, U, John, J, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
3SZE
 
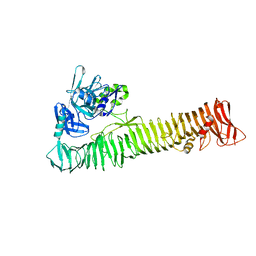 | | Crystal structure of the passenger domain of the E. coli autotransporter EspP | | Descriptor: | Serine protease espP | | Authors: | Khan, S, Mian, H.S, Sandercock, L.E, Battaile, K.P, Lam, R, Chirgadze, N.Y, Pai, E.F. | | Deposit date: | 2011-07-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Passenger Domain of the Escherichia coli Autotransporter EspP.
J.Mol.Biol., 413, 2011
|
|
2OBA
 
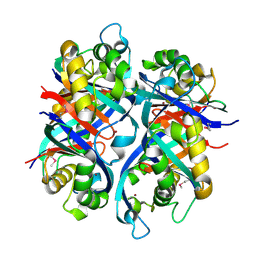 | | Pseudomonas aeruginosa 6-pyruvoyl tetrahydrobiopterin synthase | | Descriptor: | Probable 6-pyruvoyl tetrahydrobiopterin synthase, ZINC ION | | Authors: | McGrath, T.E, Kisselman, G, Battaile, K, Romanov, V, Wu-Brown, J, Guthrie, J, Virag, C, Mansoury, K, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Pseudomonas aeruginosa 6-pyruvoyl tetrahydrobiopterin synthase
TO BE PUBLISHED
|
|
4HHA
 
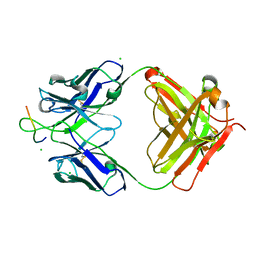 | | Anti-Human Cytomegalovirus (HCMV) Fab KE5 with epitope peptide AD-2S1 | | Descriptor: | Antibody KE5, CHLORIDE ION, Fab KE5, ... | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Pfoh, R, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-09 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4HH9
 
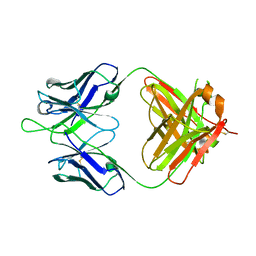 | | Anti-Human Cytomegalovirus (HCMV) Fab KE5 | | Descriptor: | Fab KE5, heavy chain, light chain | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Pfoh, R, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-09 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4HIH
 
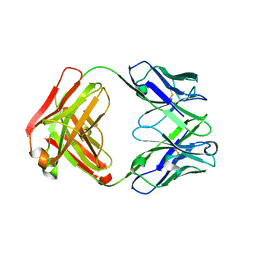 | | Anti-Streptococcus pneumoniae 23F Fab 023.102 with bound rhamnose. | | Descriptor: | Antibody 023.102, Fab 023.102, alpha-L-rhamnopyranose | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
3UNC
 
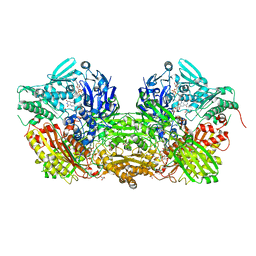 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase to 1.65A Resolution | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
3UM9
 
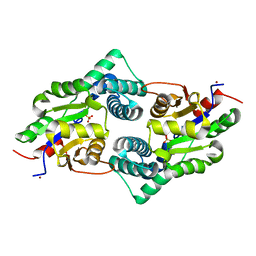 | | Crystal Structure of the Defluorinating L-2-Haloacid Dehalogenase Bpro0530 | | Descriptor: | Haloacid dehalogenase, type II, NICKEL (II) ION, ... | | Authors: | Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-12 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
3UNI
 
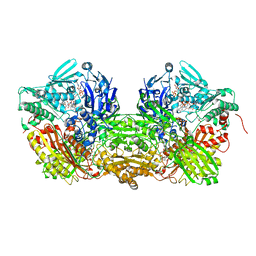 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with NADH Bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-HYDROXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
