2P8M
 
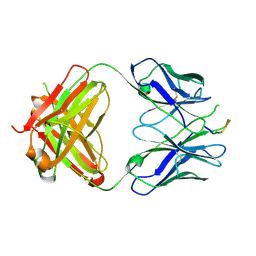 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELLELDKWASLWN in new crystal form | | Descriptor: | gp41 peptide, nmAb 2F5, heavy chain, ... | | Authors: | Julien, J.P, Bryson, S, Pai, E.F. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
2PR4
 
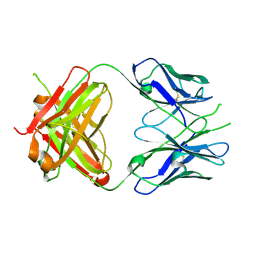 | | Crystal Structure of Fab' from the HIV-1 Neutralizing Antibody 2F5 | | Descriptor: | nmAb 2F5 Fab' Heavy Chain, nmAb 2F5 Fab' light Chain | | Authors: | Bryson, S, Julien, J.-P, Pai, E.F. | | Deposit date: | 2007-05-03 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
2Q8L
 
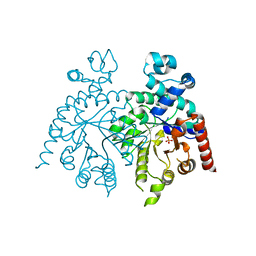 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Plasmodium falciparum | | Descriptor: | Orotidine-monophosphate-decarboxylase, PHOSPHATE ION | | Authors: | Liu, Y, Lau, W, Lew, J, Amani, M, Hui, R, Pai, E.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-11 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Plasmodium falciparum.
To be Published
|
|
1U8P
 
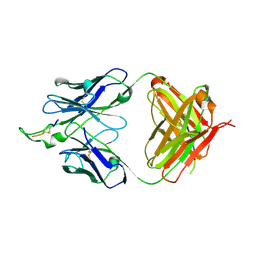 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ECDKWCS | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
3W07
 
 | |
3VSB
 
 | | SUBTILISIN CARLSBERG D-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
3NTS
 
 | |
3NUR
 
 | | Crystal structure of a putative amidohydrolase from Staphylococcus aureus | | Descriptor: | Amidohydrolase, CALCIUM ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Lam, K, Soloveychik, M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative amidohydrolase from Staphylococcus aureus
To be Published
|
|
1LOS
 
 | |
3WJW
 
 | | Wild-type orotidine 5'-monophosphate decarboxylase from M. thermoautotrophicus complexed with 6-methyl-UMP | | Descriptor: | 6-methyluridine 5'-(dihydrogen phosphate), Orotidine 5'-phosphate decarboxylase | | Authors: | Fujihashi, M, Kuroda, S, Pai, E.F, Miki, K. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Substrate distortion contributes to the catalysis of orotidine 5'-monophosphate decarboxylase.
J.Am.Chem.Soc., 135, 2013
|
|
1LP6
 
 | |
1DVJ
 
 | | CRYSTAL STRUCTURE OF OROTIDINE MONOPHOSPHATE DECARBOXYLASE COMPLEXED WITH 6-AZAUMP | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, OROTIDINE 5'-PHOSPHATE DECARBOXYLASE | | Authors: | Wu, N, Mo, Y, Gao, J, Pai, E.F. | | Deposit date: | 2000-03-30 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Electrostatic stress in catalysis: structure and mechanism of the enzyme orotidine monophosphate decarboxylase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1LOL
 
 | | Crystal structure of orotidine monophosphate decarboxylase complex with XMP | | Descriptor: | 1,3-BUTANEDIOL, XANTHOSINE-5'-MONOPHOSPHATE, orotidine 5'-monophosphate decarboxylase | | Authors: | Wu, N, Pai, E.F. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of inhibitor complexes reveal an alternate binding mode in orotidine-5'-monophosphate decarboxylase.
J.Biol.Chem., 277, 2002
|
|
3G3M
 
 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 5-fluoro-6-iodo-UMP | | Descriptor: | 5-FLUORO-URIDINE-5'-MONOPHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Bello, A.M, Poduch, E, Kotra, L.P, Pai, E.F. | | Deposit date: | 2009-02-02 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
1U8O
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKHAS | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
3WJY
 
 | |
1LVW
 
 | | Crystal structure of glucose-1-phosphate thymidylyltransferase, RmlA, complex with dTDP | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Dong, A, Christendat, D, Pai, E.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-05-29 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glucose-1-phosphate thymidylyltransferase, RmlA, complex with dTDP
To be Published
|
|
1M8K
 
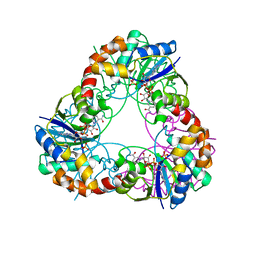 | |
1EJ2
 
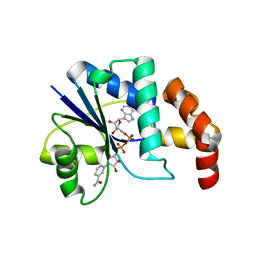 | | Crystal structure of methanobacterium thermoautotrophicum nicotinamide mononucleotide adenylyltransferase with bound NAD+ | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Saridakis, V, Christendat, D, Kimber, M.S, Edwards, A.M, Pai, E.F, Midwest Center for Structural Genomics (MCSG), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-29 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.
J.Biol.Chem., 276, 2001
|
|
3PQR
 
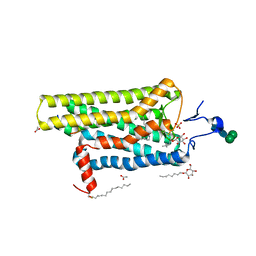 | | Crystal structure of Metarhodopsin II in complex with a C-terminal peptide derived from the Galpha subunit of transducin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, ... | | Authors: | Choe, H.-W, Kim, Y.J, Park, J.H, Morizumi, T, Pai, E.F, Krauss, N, Hofmann, K.P, Scheerer, P, Ernst, O.P. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of metarhodopsin II.
Nature, 471, 2011
|
|
3PXO
 
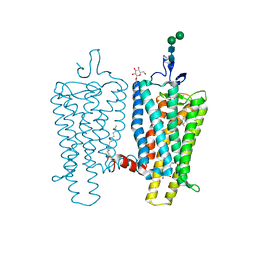 | | Crystal structure of Metarhodopsin II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, RETINAL, ... | | Authors: | Choe, H.-W, Kim, Y.J, Park, J.H, Morizumi, T, Pai, E.F, Krauss, N, Hofmann, K.P, Scheerer, P, Ernst, O.P. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of metarhodopsin II.
Nature, 471, 2011
|
|
3EYF
 
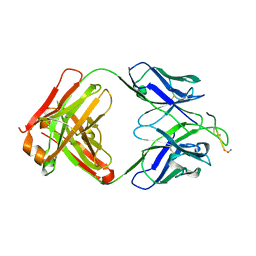 | | Crystal structure of anti-human cytomegalovirus antibody 8f9 plus gB peptide | | Descriptor: | 8f9 Fab, AD-2, GLYCEROL, ... | | Authors: | Thomson, C.A, Bryson, S, McLean, G.R, Creagh, A.L, Pai, E.F, Schrader, J.W. | | Deposit date: | 2008-10-20 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus.
Embo J., 27, 2008
|
|
3D7R
 
 | | Crystal structure of a putative esterase from Staphylococcus aureus | | Descriptor: | BROMIDE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lam, R, Battaile, K.P, Chan, T, Romanov, V, Lam, K, Soloveychik, M, Wu-Brown, J, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2008-05-21 | | Release date: | 2009-06-09 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of a putative esterase from Staphylococcus aureus
To be Published
|
|
3THQ
 
 | |
1KM2
 
 | |
