7JM2
 
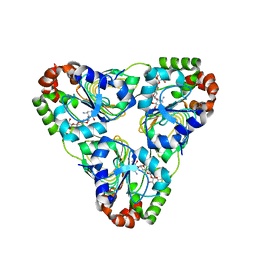 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin | | Descriptor: | APRAMYCIN, Aminocyclitol acetyltransferase ApmA, CHLORIDE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with apramycin
To Be Published
|
|
7K8M
 
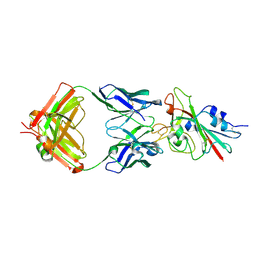 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C102 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C102 Fab Heavy Chain, C102 Fab Light Chain, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8V
 
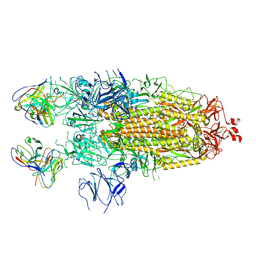 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C110 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C110 Fab Heavy Chain, ... | | Authors: | Dam, K.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7KDE
 
 | |
7K8O
 
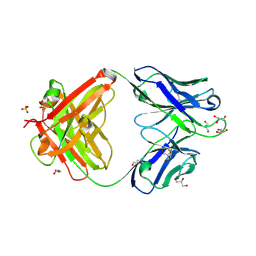 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C002 | | Descriptor: | C002 Fab Heavy Chain, C002 Fab Light Chain, GLYCEROL, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8W
 
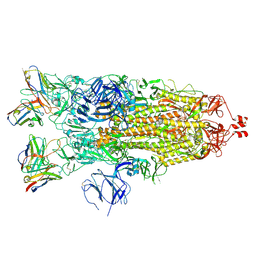 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C119 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C119 Fab Heavy Chain, ... | | Authors: | Sharaf, N.G, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
6FJO
 
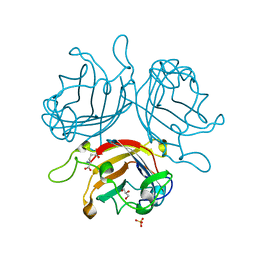 | | Adenovirus species 26 knob protein, very high resolution | | Descriptor: | Fiber, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Human adenovirus type 26 uses sialic acid-bearing glycans as a primary cell entry receptor.
Sci Adv, 5, 2019
|
|
4CQQ
 
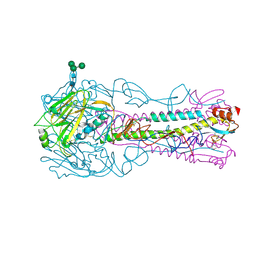 | | H5 (VN1194) Ser227Asn/Gln196Arg Mutant Haemagglutinin in Complex with Avian Receptor Analogue 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Xiong, X, Xiao, H, Martin, S.R, Coombs, P.J, Liu, J, Collins, P.J, Vachieri, S.G, Walker, P.A, Lin, Y.P, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Enhanced Human Receptor Binding by H5 Haemagglutinins.
Virology, 456, 2014
|
|
4CQY
 
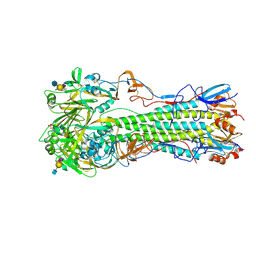 | | H5 (tyTy) Del133/Ile155Thr Mutant Haemagglutinin in Complex with Avian Receptor Analogue LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ HA1, HAEMAGGLUTININ HA2, ... | | Authors: | Xiong, X, Xiao, H, Martin, S.R, Coombs, P.J, Liu, J, Collins, P.J, Vachieri, S.G, Walker, P.A, Lin, Y.P, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enhanced Human Receptor Binding by H5 Haemagglutinins.
Virology, 456, 2014
|
|
4CQV
 
 | | Crystal structure of H5 (tyTy) Del133/Ile155Thr Mutant Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ HA1, HAEMAGGLUTININ HA2, ... | | Authors: | Xiong, X, Xiao, H, Martin, S.R, Coombs, P.J, Liu, J, Collins, P.J, Vachieri, S.G, Walker, P.A, Lin, Y.P, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Enhanced Human Receptor Binding by H5 Haemagglutinins.
Virology, 456, 2014
|
|
4CQZ
 
 | | Crystal Structure of H5 (VN1194) Gln196Arg Mutant Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, J, Xiong, X, Xiao, H, Martin, S.R, Coombs, P.J, Collins, P.J, Vachieri, S.G, Walker, P.A, Lin, Y.P, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enhanced Human Receptor Binding by H5 Haemagglutinins.
Virology, 456, 2014
|
|
4CR0
 
 | | Crystal Structure of H5 (VN1194) Asn186Lys/Gly143Arg Mutant Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2 | | Authors: | Collins, P.J, Vachieri, S.G, Xiong, X, Xiao, H, Martin, S.R, Coombs, P.J, Liu, J, Walker, P.A, Lin, Y.P, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Enhanced Human Receptor Binding by H5 Haemagglutinins.
Virology, 456, 2014
|
|
6FR3
 
 | | 003 TCR Study of CDR Loop Flexibility | | Descriptor: | 003 TCR Alpha Chain, 003 TCR Beta Chain, 1,2-ETHANEDIOL, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2018-02-15 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | In Silicoand Structural Analyses Demonstrate That Intrinsic Protein Motions Guide T Cell Receptor Complementarity Determining Region Loop Flexibility.
Front Immunol, 9, 2018
|
|
1R1N
 
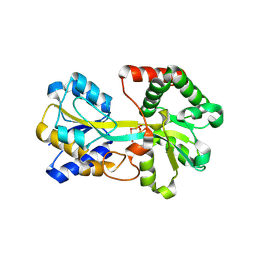 | | Tri-nuclear oxo-iron clusters in the ferric binding protein from N. gonorrhoeae | | Descriptor: | Ferric-iron Binding Protein, OXO-IRON CLUSTER 1, OXO-IRON CLUSTER 2, ... | | Authors: | Zhu, H, Alexeev, D, Hunter, D.J, Campopiano, D.J, Sadler, P.J. | | Deposit date: | 2003-09-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Oxo-iron clusters in a bacterial iron-trafficking protein: new roles for a conserved motif.
Biochem.J., 376, 2003
|
|
6FR4
 
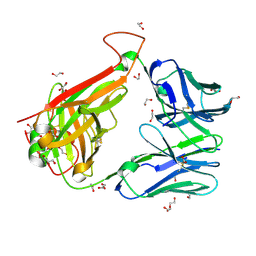 | | 003 TCR Study of CDR Loop Flexibility | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2018-02-15 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | In Silicoand Structural Analyses Demonstrate That Intrinsic Protein Motions Guide T Cell Receptor Complementarity Determining Region Loop Flexibility.
Front Immunol, 9, 2018
|
|
1S1S
 
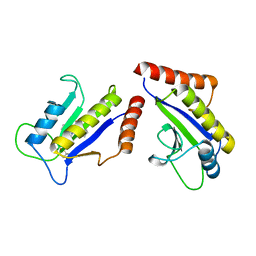 | | Crystal Structure of ZipA in complex with indoloquinolizin 10b | | Descriptor: | Cell division protein zipA, N-{3-[(12bS)-7-oxo-1,3,4,6,7,12b-hexahydroindolo[2,3-a]quinolizin-12(2H)-yl]propyl}propane-2-sulfonamide | | Authors: | Jennings, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenny, C.H, Moghazeh, S.L, Petersen, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | Deposit date: | 2004-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
1SDQ
 
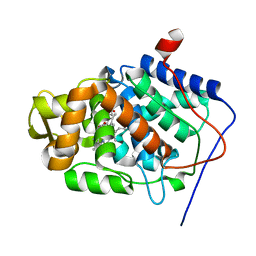 | | Structure of reduced-NO adduct of mesopone cytochrome c peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, FE-(4-MESOPORPHYRINONE)-R-ISOMER, ... | | Authors: | Bhaskar, B, Immoos, C.E, Sulc, F, Cohem, M.S, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2004-02-13 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structures of resting (Fe3+), reduced (Fe2+) and NO-bound states of mesopone cytochrome c peroxidase (MpCcP) (R-isomer)
To be Published
|
|
4BH4
 
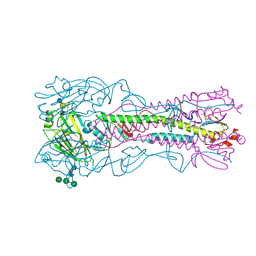 | | Haemagglutinin from a Transmissible Mutant H5 Influenza Virus in Complex with Avian Receptor Analogue 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
4BH2
 
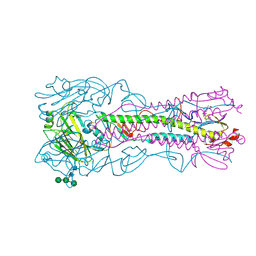 | | Crystal Structure of the Haemagglutinin from a Transmissible Mutant H5 Influenza Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
1SJE
 
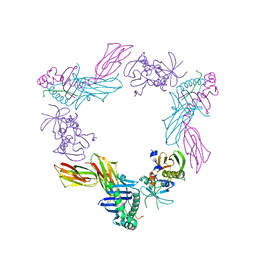 | | HLA-DR1 complexed with a 16 residue HIV capsid peptide bound in a hairpin conformation | | Descriptor: | Enterotoxin type C-3, GAG polyprotein, HLA class II histocompatibility antigen, ... | | Authors: | Zavala-Ruiz, Z, Strug, I, Walker, B.D, Norris, P.J, Stern, L.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A hairpin turn in a class II MHC-bound peptide orients residues outside the binding groove for T cell recognition.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4BH0
 
 | | H5 (tyTy) Influenza Virus Haemagglutinin in Complex with Human Receptor Analogue 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
1T3P
 
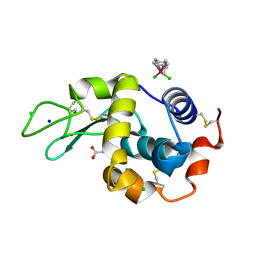 | | Half-sandwich arene ruthenium(II)-enzyme complex | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | McNae, I.W, Fishburne, K, Habtemariam, A, Hunter, T.M, Melchart, M, Wang, F, Walkinshaw, M.D, Sadler, P.J. | | Deposit date: | 2004-04-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Half-sandwich arene ruthenium(II)-enzyme complex
CHEM.COMMUN.(CAMB.), 16, 2004
|
|
4RP2
 
 | |
8V2Z
 
 | | Cryo-EM Structure of Smooth Muscle Gamma Actin (ACTG2) Mutant R257C | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, gamma-enteric smooth muscle, ... | | Authors: | Palmer, N.J, Carman, P.J, Ceron, R.H, Dominguez, R. | | Deposit date: | 2023-11-25 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Smooth Muscle Gamma Actin (ACTG2) Filament Mutant R257C
To Be Published
|
|
1R4X
 
 | | Crystal Structure Analys of the Gamma-COPI Appendage domain | | Descriptor: | Coatomer gamma subunit, MAGNESIUM ION | | Authors: | Watson, P.J, Frigerio, G, Collins, B.M, Duden, R, Owen, D.J. | | Deposit date: | 2003-10-09 | | Release date: | 2003-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gamma-COP appendage domain - structure and function
Traffic, 5, 2004
|
|
