8F2O
 
 | |
8F2N
 
 | | Phi-29 partially-expanded fiberless prohead | | Descriptor: | Major capsid protein | | Authors: | Woodson, M.E, Morais, M.C, Scott, S.D, Choi, K.H, Jardine, P.J, Zhang, W. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Phi-29 partially-expanded fiberless prohead
To Be Published
|
|
8F2M
 
 | |
7QFU
 
 | | Crystal Structure of AtlA catalytic domain from Enterococcus feacalis | | Descriptor: | GLYCEROL, Peptidoglycan hydrolase | | Authors: | Zamboni, V, Barelier, S, Dixon, R, Galley, N, Ghanem, A, Cahuzac, H, Salamaga, B, Davis, P.J, Mesnage, S, Vincent, F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for substrate recognition and septum cleavage by AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis.
J.Biol.Chem., 298, 2022
|
|
8EJV
 
 | | The crystal structure of Pseudomonas putida PcaR in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR in complex with succinate
To Be Published
|
|
8EJU
 
 | | The crystal structure of Pseudomonas putida PcaR | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Transcription regulatory protein (Pca regulon), ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR
To Be Published
|
|
1S1J
 
 | | Crystal Structure of ZipA in complex with indoloquinolizin inhibitor 1 | | Descriptor: | (12bS)-1,2,3,4,12,12b-hexahydroindolo[2,3-a]quinolizin-7(6H)-one, Cell division protein zipA | | Authors: | Jenning, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenney, C.H, Moghazeh, S.L, Peterson, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | Deposit date: | 2004-01-06 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1S9G
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | Descriptor: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
8FNW
 
 | | Structure of RdrA-RdrB complex from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, Archaeal ATPase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
2CKS
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THERMOBIFIDA FUSCA ENDOGLUCANASE CEL5A (E5) | | Descriptor: | BENZAMIDINE, ENDOGLUCANASE E-5, SODIUM ION, ... | | Authors: | Berglund, G.I, Gualfetti, P.J, Requadt, C, Gross, L.S, Bergfors, T, Shaw, A, Saldajeno, M, Mitchinson, C, Sandgren, M. | | Deposit date: | 2006-04-21 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Catalytic Domain of Thermobifida Fusca Endoglucanase Cel5A in Complex with Cellotetraose
To be Published
|
|
8FNT
 
 | | Structure of RdrA from Escherichia coli RADAR defense system | | Descriptor: | Archaeal ATPase | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FLP
 
 | | NMR Solution Structure of LvIC analogue | | Descriptor: | Alpha-conotoxin LvIC analogue | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | Discovery, Characterization, and Engineering of LvIC, an alpha 4/4-Conotoxin That Selectively Blocks Rat alpha 6/ alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 66, 2023
|
|
8FNV
 
 | | Structure of RdrB from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNU
 
 | | Structure of RdrA from Streptococcus suis RADAR defense system | | Descriptor: | KAP NTPase domain-containing protein | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
2CMB
 
 | | Structural Basis for Inhibition of Protein Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics | | Descriptor: | N-{[4-(1,1-DIOXIDO-3-OXO-2,3-DIHYDROISOTHIAZOL-5-YL)PHENYL]ACETYL}-L-PHENYLALANYL-4-(1,1-DIOXIDO-3-OXO-2,3-DIHYDROISOTHIAZOL-5-YL)-L-PHENYLALANINAMIDE, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1, octyl beta-D-glucopyranoside | | Authors: | Ala, P.J, Gonneville, L, Hillman, M.C, Becker-Pasha, M, Wei, M, Reid, B.G, Klabe, R, Yue, E.W, Wayland, B, Douty, B, Combs, A.P, Polam, P, Wasserman, Z, Bower, M, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-04 | | Release date: | 2006-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics.
J.Biol.Chem., 281, 2006
|
|
2BOB
 
 | | Potassium channel KcsA-Fab complex in thallium with tetrabutylammonium (TBA) | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, COBALT (II) ION, ... | | Authors: | Lenaeus, M.J, Vamvouka, M, Focia, P.J, Gross, A. | | Deposit date: | 2005-04-09 | | Release date: | 2005-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Basis of Tea Blockade in a Model Potassium Channel
Nat.Struct.Mol.Biol., 12, 2005
|
|
8F8Q
 
 | | Cryo-EM structure of the CapZ-capped barbed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
8F8P
 
 | | Cryo-EM structure of F-actin in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
8F8R
 
 | | Cryo-EM structure of the free barbed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
7QCP
 
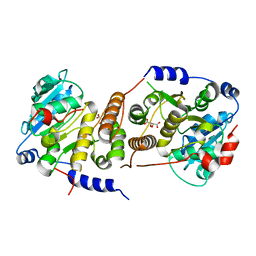 | |
8F8T
 
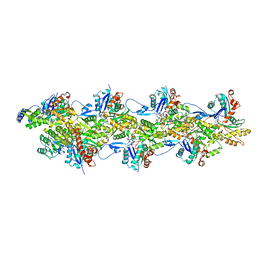 | | Cryo-EM structure of the Tropomodulin-capped pointed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
8F8S
 
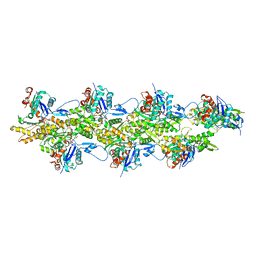 | | Cryo-EM structure of the free pointed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
2CNW
 
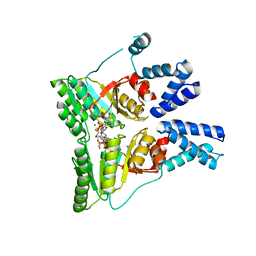 | | GDPALF4 complex of the SRP GTPases Ffh and FtsY | | Descriptor: | CELL DIVISION PROTEIN FTSY, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Focia, P.J, Gawronski-Salerno, J, Coon V, J.S, Freymann, D.M. | | Deposit date: | 2006-05-24 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of a Gdp:Alf(4) Complex of the Srp Gtpases Ffh and Ftsy, and Identification of a Peripheral Nucleotide Interaction Site.
J.Mol.Biol., 360, 2006
|
|
1ONY
 
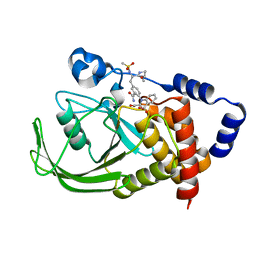 | | Oxalyl-Aryl-Amino Benzoic Acid inhibitors of PTP1B, compound 17 | | Descriptor: | 2-{[2-(2-CARBAMOYL-VINYL)-4-(2-METHANESULFONYLAMINO-2-PENTYLCARBAMOYL-ETHYL)-PHENYL]-OXALYL-AMINO}-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Szczepankiewicz, B.G, Pei, Z, Janowich, D.A, Xin, Z, Hadjuk, P.J, Abad-Zapatero, C, Liang, H, Hutchins, C.W, Fesik, S.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Mika, A.K, Zinker, B.A, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and Structure-Activity Relationship of Oxalylarylaminobenzoic
Acids as Inhibitors of Protein Tyrosine Phosphatase 1B
J.Med.Chem., 46, 2003
|
|
7QIB
 
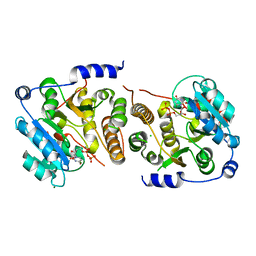 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UDP | | Descriptor: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MAGNESIUM ION, ... | | Authors: | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 8.5 in complex with UDP
To Be Published
|
|
