6PIO
 
 | |
6PIN
 
 | |
6PTA
 
 | | Crystal structure of the ARF family small GTPase ARF1 from Candida albicans in complex with GDP | | Descriptor: | ADP-ribosylation factor, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-15 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the ARF family small GTPase ARF1 from Candida albicans in complex with GDP
To Be Published
|
|
3KM1
 
 | | ZINC-Reconstituted TOMATO CHLOROPLAST SUPEROXIDE DISMUTASE | | Descriptor: | GLYCEROL, Superoxide dismutase [Cu-Zn], chloroplastic, ... | | Authors: | Galaleldeen, A, Taylor, A.B, Thomas, S.T, Hart, P.J. | | Deposit date: | 2009-11-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biophysical properties of tomato chloroplast sod
To be Published
|
|
1ZUW
 
 | | Crystal structure of B.subtilis glutamate racemase (RacE) with D-Glu | | Descriptor: | D-GLUTAMIC ACID, glutamate racemase 1 | | Authors: | Ruzheinikov, S.N, Taal, M.A, Sedelnikova, S.E, Baker, P.J, Rice, D.W. | | Deposit date: | 2005-06-01 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-Induced Conformational Changes in Bacillus subtilis Glutamate Racemase and Their Implications for Drug Discovery
Structure, 13, 2005
|
|
5HZM
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN ATOM OR ION | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, MIN, J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-02 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
5HMN
 
 | | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol | | Descriptor: | AAC3-I, COENZYME A, TETRAETHYLENE GLYCOL | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol
To Be Published
|
|
1YAZ
 
 | | AZIDE-BOUND YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | AZIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
4LTA
 
 | |
5HYJ
 
 | | 1E6 TCR in Complex with HLA-A02 carrying AQWGPDPAAA | | Descriptor: | ALA-GLN-TRP-GLY-PRO-ASP-PRO-ALA-ALA-ALA, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2016-02-01 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A "hotspot" for autoimmune T cells in type 1 diabetes.
J.Clin.Invest., 126, 2016
|
|
1ZCY
 
 | | apo form of a mutant of glycogenin in which Asp159 is replaced by Ser | | Descriptor: | Glycogenin-1, SULFATE ION | | Authors: | Hurley, T.D, Stout, S.L, Miner, E, Zhou, J, Roach, P.J. | | Deposit date: | 2005-04-13 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Requirements for catalysis in mammalian glycogenin.
J.Biol.Chem., 280, 2005
|
|
1ZCU
 
 | | apo form of the 162S mutant of glycogenin | | Descriptor: | Glycogenin-1 | | Authors: | Hurley, T.D, Stout, S.L, Miner, E, Zhou, J, Roach, P.J. | | Deposit date: | 2005-04-13 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Requirements for catalysis in mammalian glycogenin.
J.Biol.Chem., 280, 2005
|
|
4BH1
 
 | | H5 (tyTy) Influenza Virus Haemagglutinin in Complex with Avian Receptor Analogue 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
4BH3
 
 | | Haemagglutinin from a Transmissible Mutant H5 Influenza Virus in Complex with Human Receptor Analogue 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
4BH4
 
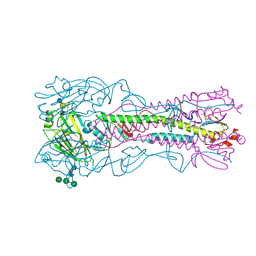 | | Haemagglutinin from a Transmissible Mutant H5 Influenza Virus in Complex with Avian Receptor Analogue 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
4BH2
 
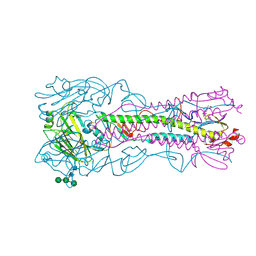 | | Crystal Structure of the Haemagglutinin from a Transmissible Mutant H5 Influenza Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Coombs, P.J, Martin, S.R, Liu, J, Xiao, H, McCauley, J.W, Locher, K, Walker, P.A, Collins, P.J, Kawaoka, Y, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-03-29 | | Release date: | 2013-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus
Nature, 497, 2013
|
|
1Y7J
 
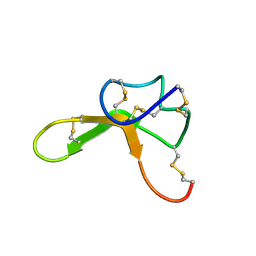 | | NMR structure family of Human Agouti Signalling Protein (80-132: Q115Y, S124Y) | | Descriptor: | Agouti Signaling Protein | | Authors: | McNulty, J.C, Jackson, P.J, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Dawson, P.E, Millhauser, G.L. | | Deposit date: | 2004-12-08 | | Release date: | 2005-02-15 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structures of the agouti signaling protein.
J.Mol.Biol., 346, 2005
|
|
1T80
 
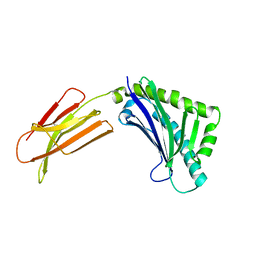 | | Zn-alpha-2-glycoprotein; CHO-ZAG PEG 200 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Zinc-alpha-2-glycoprotein | | Authors: | Delker, S.L, West Jr, A.P, McDermott, L, Kennedy, M.W, Bjorkman, P.J. | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies of ligand binding by Zn-alpha2-glycoprotein.
J.Struct.Biol., 148, 2004
|
|
1Y7K
 
 | | NMR structure family of Human Agouti Signalling Protein (80-132: Q115Y, S124Y) | | Descriptor: | Agouti Signaling Protein | | Authors: | McNulty, J.C, Jackson, P.J, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Dawson, P.E, Millhauser, G.L. | | Deposit date: | 2004-12-08 | | Release date: | 2005-02-15 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structures of the agouti signaling protein.
J.Mol.Biol., 346, 2005
|
|
5IR0
 
 | | Crystal structure of protein of unknown function ORF19 from Vibrio cholerae O1 PICI-like element, C57S I109M mutant | | Descriptor: | CITRIC ACID, Uncharacterized protein ORF19 | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (3.297 Å) | | Cite: | Crystal structure of protein of unknown function ORF19 from Vibrio cholerae O1 PICI-like element, C57S I109M mutant
To Be Published
|
|
1T7W
 
 | | Zn-alpha-2-glycoprotein; CHO-ZAG PEG 400 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Zinc-alpha-2-glycoprotein | | Authors: | Delker, S.L, West Jr, A.P, McDermott, L, Kennedy, M.W, Bjorkman, P.J. | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of ligand binding by Zn-alpha2-glycoprotein.
J.Struct.Biol., 148, 2004
|
|
1T7X
 
 | | Zn-alpha-2-glycoprotein; refolded CHO-ZAG PEG 400 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Zinc-alpha-2-glycoprotein | | Authors: | Delker, S.L, West Jr, A.P, McDermott, L, Kennedy, M.W, Bjorkman, P.J. | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystallographic studies of ligand binding by Zn-alpha2-glycoprotein.
J.Struct.Biol., 148, 2004
|
|
2B5W
 
 | | Crystal structure of D38C glucose dehydrogenase mutant from Haloferax mediterranei | | Descriptor: | CITRATE ANION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Britton, K.L, Baker, P.J, Fisher, M, Ruzheinikov, S, Gilmour, D.J, Bonete, M.-J, Ferrer, J, Pire, C, Esclapez, J, Rice, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1A14
 
 | | COMPLEX BETWEEN NC10 ANTI-INFLUENZA VIRUS NEURAMINIDASE SINGLE CHAIN ANTIBODY WITH A 5 RESIDUE LINKER AND INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NC10 FV (HEAVY CHAIN), ... | | Authors: | Malby, R.L, Mccoy, A.J, Kortt, A.A, Hudson, P.J, Colman, P.M. | | Deposit date: | 1997-12-21 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of single-chain Fv-neuraminidase complexes.
J.Mol.Biol., 279, 1998
|
|
1B4T
 
 | | H48C YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 1999-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
