1F81
 
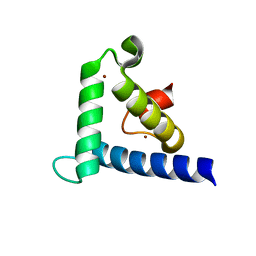 | | SOLUTION STRUCTURE OF THE TAZ2 DOMAIN OF THE TRANSCRIPTIONAL ADAPTOR PROTEIN CBP | | Descriptor: | CREB-BINDING PROTEIN, ZINC ION | | Authors: | De Guzman, R.N, Liu, H.L, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2000-06-28 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP.
J.Mol.Biol., 303, 2000
|
|
2AGH
 
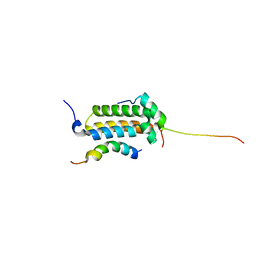 | | Structural basis for cooperative transcription factor binding to the CBP coactivator | | Descriptor: | Crebbp protein, Myb proto-oncogene protein, Zinc finger protein HRX | | Authors: | De Guzman, R.N, Goto, N.K, Dyson, H.J, Wright, P.E. | | Deposit date: | 2005-07-26 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Cooperative Transcription Factor Binding to the CBP Coactivator
J.Mol.Biol., 355, 2006
|
|
3QL0
 
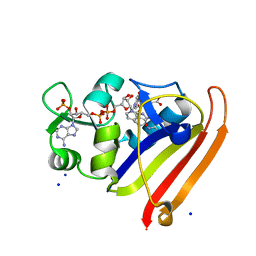 | | Crystal structure of N23PP/S148A mutant of E. coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
3QU3
 
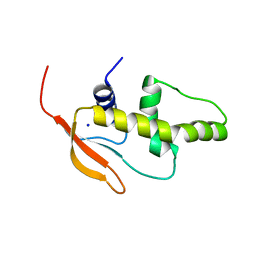 | | Crystal structure of IRF-7 DBD apo form | | Descriptor: | 1,2-ETHANEDIOL, Interferon regulatory factor 7, SODIUM ION | | Authors: | De Ioannes, P.E, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2011-02-23 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of apo IRF-3 and IRF-7 DNA binding domains: effect of loop L1 on DNA binding.
Nucleic Acids Res., 39, 2011
|
|
1HWY
 
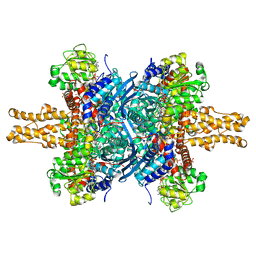 | | BOVINE GLUTAMATE DEHYDROGENASE COMPLEXED WITH NAD AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, GLUTAMATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Smith, T.J, Peterson, P.E, Schmidt, T, Fang, J, Stanley, C.A. | | Deposit date: | 2001-01-10 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of bovine glutamate dehydrogenase complexes elucidate the mechanism of purine regulation.
J.Mol.Biol., 307, 2001
|
|
3QU6
 
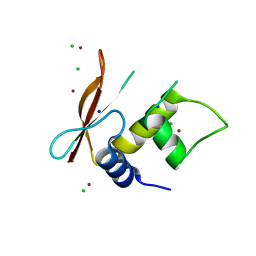 | | Crystal structure of IRF-3 DBD free form | | Descriptor: | CHLORIDE ION, IRF3 protein, SODIUM ION, ... | | Authors: | De Ioannes, P.E, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2011-02-23 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of apo IRF-3 and IRF-7 DNA binding domains: effect of loop L1 on DNA binding.
Nucleic Acids Res., 39, 2011
|
|
3OK0
 
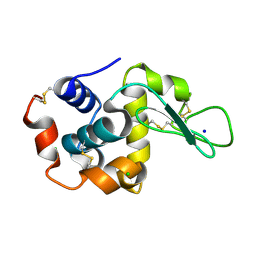 | | E35A Mutant of Hen Egg White Lysozyme (HEWL) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | O'Meara, F, Bradley, J, O'Rourke, P.E, Webb, H, Tynan-Connolly, B.M, Nielsen, J.E. | | Deposit date: | 2010-08-24 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | E35A Mutant of Hen Egg White Lysozyme (HEWL)
TO BE PUBLISHED
|
|
3OJP
 
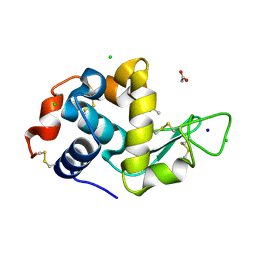 | | D52N Mutant of Hen Egg White Lysozyme (HEWL) | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | O'Meara, F, Bradley, J, O'Rourke, P.E, Webb, H, Tynan-Connolly, B.M, Nielsen, J.E. | | Deposit date: | 2010-08-23 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | D52N Mutant of Hen Egg White Lysozyme (HEWL)
TO BE PUBLISHED
|
|
1FAS
 
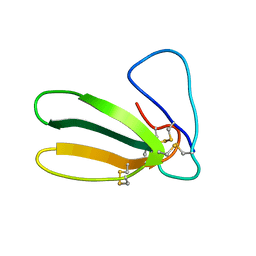 | | 1.9 ANGSTROM RESOLUTION STRUCTURE OF FASCICULIN 1, AN ANTI-ACETYLCHOLINESTERASE TOXIN FROM GREEN MAMBA SNAKE VENOM | | Descriptor: | FASCICULIN 1 | | Authors: | Le Du, M.H, Marchot, P, Bougis, P.E, Fontecilla-Camps, J.C. | | Deposit date: | 1992-08-07 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.9-A resolution structure of fasciculin 1, an anti-acetylcholinesterase toxin from green mamba snake venom.
J.Biol.Chem., 267, 1992
|
|
3QL3
 
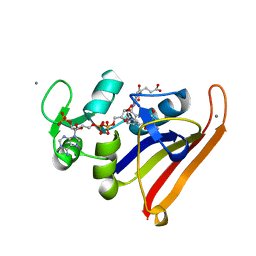 | | Re-refined coordinates for PDB entry 1RX2 | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
1G7O
 
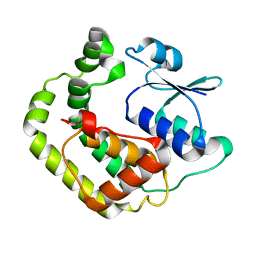 | | NMR SOLUTION STRUCTURE OF REDUCED E. COLI GLUTAREDOXIN 2 | | Descriptor: | GLUTAREDOXIN 2 | | Authors: | Xia, B, Vlamis-Gardikas, A, Holmgren, A, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-11-10 | | Release date: | 2001-07-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Escherichia coli glutaredoxin-2 shows similarity to mammalian glutathione-S-transferases.
J.Mol.Biol., 310, 2001
|
|
1ZNF
 
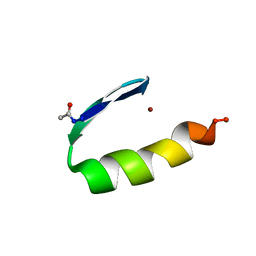 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SINGLE ZINC FINGER DNA-BINDING DOMAIN | | Descriptor: | 31ST ZINC FINGER FROM XFIN, ZINC ION | | Authors: | Lee, M.S, Gippert, G.P, Soman, K.V, Case, D.A, Wright, P.E. | | Deposit date: | 1989-09-25 | | Release date: | 1991-07-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of a single zinc finger DNA-binding domain.
Science, 245, 1989
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
2AVX
 
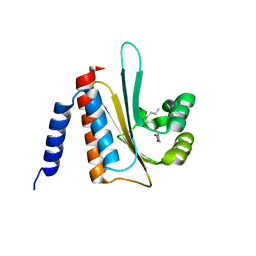 | | solution structure of E coli SdiA1-171 | | Descriptor: | N-(2-OXOTETRAHYDROFURAN-3-YL)OCTANAMIDE, Regulatory protein sdiA | | Authors: | Yao, Y, Martinez-Yamout, M.A, Dickerson, T.J, Brogan, A.P, Wright, P.E, Dyson, H.J. | | Deposit date: | 2005-08-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones.
J.Mol.Biol., 355, 2006
|
|
1JVQ
 
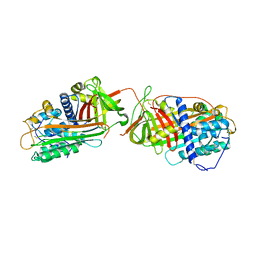 | | Crystal structure at 2.6A of the ternary complex between antithrombin, a P14-P8 reactive loop peptide, and an exogenous tetrapeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | Zhou, A, Huntington, J.A, Lomas, D.A, Carrell, R.W, Stein, P.E. | | Deposit date: | 2001-08-31 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How small peptides block and reverse serpin polymerisation
J.Mol.Biol., 342, 2004
|
|
1JAW
 
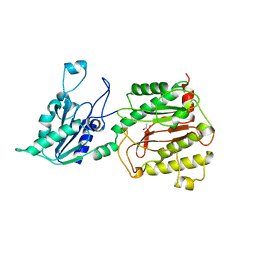 | | AMINOPEPTIDASE P FROM E. COLI LOW PH FORM | | Descriptor: | ACETATE ION, AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Wilce, M.C.J, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1LJ5
 
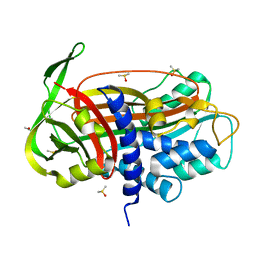 | |
7FSR
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z54615640 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSV
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z1454310449 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FT9
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z45636695 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(METHYLSULFONYL)AMINO]BENZOIC ACID, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FTA
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z228589380 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(pyrimidin-2-yl)amino]benzoic acid, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSU
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z729726784 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-N-(3-methyl-1,2,4-oxadiazol-5-yl)acetamide, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSJ
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z57744712 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FTB
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z1198269757 | | Descriptor: | 1,2-ETHANEDIOL, D-GLUTAMIC ACID, GLYCINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FST
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z26968795 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
