1VCA
 
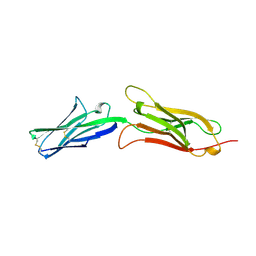 | | CRYSTAL STRUCTURE OF AN INTEGRIN-BINDING FRAGMENT OF VASCULAR CELL ADHESION MOLECULE-1 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN VASCULAR CELL ADHESION MOLECULE-1 | | Authors: | Jones, E.Y, Harlos, K, Bottomley, M.J, Robinson, R.C, Driscoll, P.C, Edwards, R.M, Clements, J.M, Dudgeon, T.J, Stuart, D.I. | | Deposit date: | 1995-03-21 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 A resolution.
Nature, 373, 1995
|
|
1PNK
 
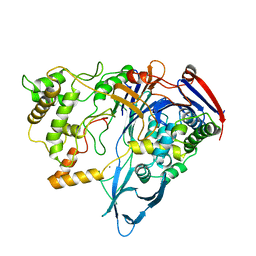 | |
1PNJ
 
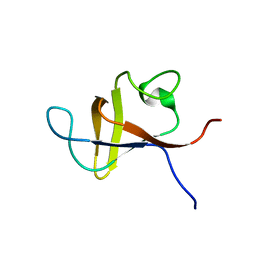 | | SOLUTION STRUCTURE AND LIGAND-BINDING SITE OF THE SH3 DOMAIN OF THE P85ALPHA SUBUNIT OF PHOSPHATIDYLINOSITOL 3-KINASE | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Booker, G.W, Gout, I, Downing, A.K, Driscoll, P.C, Boyd, J, Waterfield, M.D, Campbell, I.D. | | Deposit date: | 1993-07-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase.
Cell(Cambridge,Mass.), 73, 1993
|
|
1PNM
 
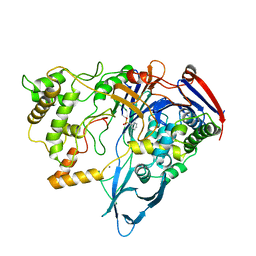 | |
3BEG
 
 | | Crystal structure of SR protein kinase 1 complexed to its substrate ASF/SF2 | | Descriptor: | ALANINE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PHOSPHOSERINE, ... | | Authors: | Ngo, J.C, Giang, K, Chakrabarti, S, Ma, C.-T, Huynh, N, Hagopian, J, Dorrestein, P.C, Fu, X.-D, Adams, J.A, Ghosh, G. | | Deposit date: | 2007-11-18 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A sliding docking interaction is essential for sequential and
processive phosphorylation of an SR protein by SRPK1
Mol.Cell, 29, 2008
|
|
1PLB
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED PARSLEY PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bagby, S, Driscoll, P.C, Harvey, T.S, Hill, H.A.O. | | Deposit date: | 1994-05-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced parsley plastocyanin.
Biochemistry, 33, 1994
|
|
1PTS
 
 | |
1STP
 
 | |
1TQQ
 
 | | Structure of TolC in complex with hexamminecobalt | | Descriptor: | COBALT HEXAMMINE(III), Outer membrane protein tolC | | Authors: | Higgins, M.K, Eswaran, J, Edwards, P.C, Schertler, G.F, Hughes, C, Koronakis, V. | | Deposit date: | 2004-06-18 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the ligand-blocked periplasmic entrance of the bacterial multidrug effllux protein TolC
J.Mol.Biol., 342, 2004
|
|
1V0H
 
 | | ASCOBATE PEROXIDASE FROM SOYBEAN CYTOSOL IN COMPLEX WITH SALICYLHYDROXAMIC ACID | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SALICYLHYDROXAMIC ACID, ... | | Authors: | Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2004-03-29 | | Release date: | 2004-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Ascorbate Peroxidase-Salicylhydroxamic Acid Complex
Biochemistry, 43, 2004
|
|
1NHN
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1NHM
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
NUCLEIC ACIDS MOL.BIOL., 9, 1995
|
|
1SFE
 
 | | ADA O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | ADA O6-METHYLGUANINE-DNA METHYLTRANSFERASE | | Authors: | Moore, M.H, Gulbis, J.M, Dodson, E.J, Demple, B, Moody, P.C.E. | | Deposit date: | 1996-06-21 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a suicidal DNA repair protein: the Ada O6-methylguanine-DNA methyltransferase from E. coli.
EMBO J., 13, 1994
|
|
1SHZ
 
 | | Crystal Structure of the p115RhoGEF rgRGS Domain in A Complex with Galpha(13):Galpha(i1) Chimera | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine Nucleotide-Binding Protein Galpha(13):Galpha(i1) Chimera, MAGNESIUM ION, ... | | Authors: | Chen, Z, Singer, W.D, Sternweis, P.C, Sprang, S.R. | | Deposit date: | 2004-02-26 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the p115RhoGEF rgRGS domain-Galpha13/i1 chimera complex suggests convergent evolution of a GTPase activator.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1QWS
 
 | | Structure of the D181N variant of catalase HPII from E. coli | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-09-03 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
1MUC
 
 | | STRUCTURE OF MUCONATE LACTONIZING ENZYME AT 1.85 ANGSTROMS RESOLUTION | | Descriptor: | MANGANESE (II) ION, MUCONATE LACTONIZING ENZYME | | Authors: | Helin, S, Kahn, P.C, Guha, B.H.L, Mallows, D.J, Goldman, A. | | Deposit date: | 1995-09-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 A resolution.
J.Mol.Biol., 254, 1995
|
|
1N4D
 
 | |
1QF7
 
 | |
1QBQ
 
 | | STRUCTURE OF RAT FARNESYL PROTEIN TRANSFERASE COMPLEXED WITH A CVIM PEPTIDE AND ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID. | | Descriptor: | ACETATE ION, ACETYL-CYS-VAL-ILE-SELENOMET-COOH PEPTIDE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, ... | | Authors: | Strickland, C.L, Windsor, W.T, Syto, R, Wang, L, Bond, R, Wu, Z, Schwartz, J, Le, H.V, Beese, L.S, Weber, P.C. | | Deposit date: | 1999-04-27 | | Release date: | 1999-06-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue
Biochemistry, 37, 1998
|
|
1QWL
 
 | | Structure of Helicobacter pylori catalase | | Descriptor: | AZIDE ION, KatA catalase, OXYGEN MOLECULE, ... | | Authors: | Loewen, P.C, Carpena, X, Perez-Luque, R, Rovira, C, Haas, R, Obenbreit, S, Nicholls, P, Fita, I. | | Deposit date: | 2003-09-02 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Helicobacter pylori Catalase, with and without Formic Acid Bound, at 1.6 A Resolution
Biochemistry, 43, 2004
|
|
1QNT
 
 | |
1TPM
 
 | | SOLUTION STRUCTURE OF THE FIBRIN BINDING FINGER DOMAIN OF TISSUE-TYPE PLASMINOGEN ACTIVATOR DETERMINED BY 1H NUCLEAR MAGNETIC RESONANCE | | Descriptor: | TISSUE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Downing, A.K, Driscoll, P.C, Harvey, T.S, Dudgeon, T.J, Smith, B.O, Baron, M, Campbell, I.D. | | Deposit date: | 1993-05-26 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fibrin binding finger domain of tissue-type plasminogen activator determined by 1H nuclear magnetic resonance.
J.Mol.Biol., 225, 1992
|
|
1U2L
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (P1) | | Descriptor: | Peroxidase/catalase HPI | | Authors: | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | Deposit date: | 2004-07-19 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TYG
 
 | | Structure of the thiazole synthase/ThiS complex | | Descriptor: | PHOSPHATE ION, Thiazole biosynthesis protein thiG, yjbS | | Authors: | Settembre, E.C, Dorrestein, P.C, Zhai, H, Chatterjee, A, McLafferty, F.W, Begley, T.P, Ealick, S.E. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Thiamin Biosynthesis in Bacillus subtilis: Structure of the Thiazole Synthase/Sulfur Carrier Protein Complex
Biochemistry, 43, 2004
|
|
1VYS
 
 | | STRUCTURE OF PENTAERYTHRITOL TETRANITRATE REDUCTASE W102Y MUTANT AND COMPLEXED WITH PICRIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-05 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
