1N4A
 
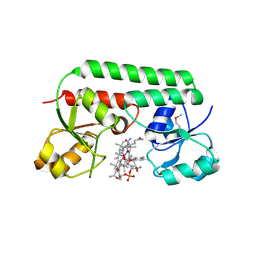 | | The Ligand Bound Structure of E.coli BtuF, the Periplasmic Binding Protein for Vitamin B12 | | Descriptor: | CYANOCOBALAMIN, Vitamin B12 transport protein btuF | | Authors: | Karpowich, N.K, Smith, P.C, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-30 | | Release date: | 2003-03-11 | | Last modified: | 2021-08-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding.
J.Biol.Chem., 278, 2003
|
|
7BEX
 
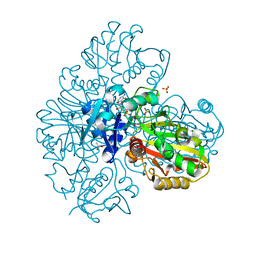 | |
7BEW
 
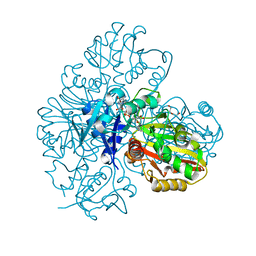 | |
1SQM
 
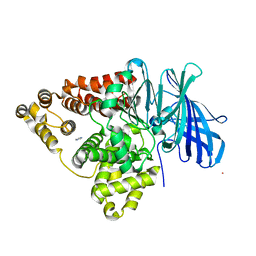 | | STRUCTURE OF [R563A] LEUKOTRIENE A4 HYDROLASE | | Descriptor: | ACETIC ACID, IMIDAZOLE, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Tholander, F.O.T, Rudberg, P.C, Thunnissen, M.M.G.M, Haeggstrom, J.Z. | | Deposit date: | 2004-03-19 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Leukotriene A4 hydrolase: identification of a common carboxylate recognition site for the epoxide hydrolase and aminopeptidase substrates
J.Biol.Chem., 279, 2004
|
|
1SRH
 
 | |
1SRG
 
 | |
1SRI
 
 | |
1SRF
 
 | |
1SRE
 
 | |
2YK5
 
 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CMP. | | Descriptor: | 1,2-ETHANEDIOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
1AEX
 
 | | STAPHYLOCOCCAL NUCLEASE, METHANE THIOL DISULFIDE TO V23C VARIANT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1997-03-01 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mobile unnatural amino acid side chains in the core of staphylococcal nuclease.
Protein Sci., 5, 1996
|
|
1QLW
 
 | |
2YFQ
 
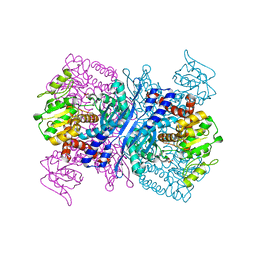 | | Crystal structure of Glutamate dehydrogenase from Peptoniphilus asaccharolyticus | | Descriptor: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE, SULFATE ION | | Authors: | Oliveira, T, Panjikar, S, Carrigan, J.B, Sharkey, M.A, Hamza, M, Engel, P.C, Khan, A.R. | | Deposit date: | 2011-04-07 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal Structure of Nad-Dependent Peptoniphilus Asaccharolyticus Glutamate Dehydrogenase Reveals Determinants of Cofactor Specificity.
J.Struct.Biol., 177, 2012
|
|
1A2T
 
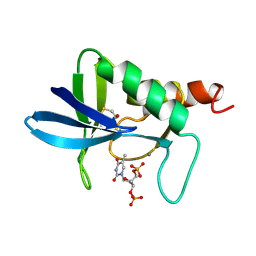 | | STAPHYLOCOCCAL NUCLEASE, B-MERCAPTOETHANOL DISULFIDE TO V23C VARIANT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1998-01-11 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mobile unnatural amino acid side chains in the core of staphylococcal nuclease.
Protein Sci., 5, 1996
|
|
1ANV
 
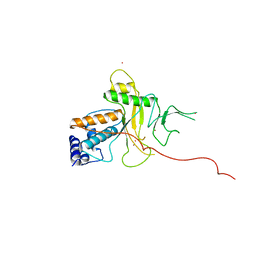 | | ADENOVIRUS 5 DBP/URANYL FLUORIDE SOAK | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, URANYL (VI) ION, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1996-03-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change of the adenovirus DNA-binding protein induced by soaking crystals with K3UO2F5 solutions.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1OKF
 
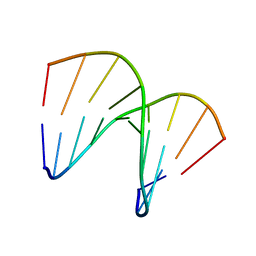 | | NMR structure of an alpha-L-LNA:RNA hybrid | | Descriptor: | 5'-D(*CP*ATLP*GP*AP*ATLP*AP*ATLP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*AP*UP*CP*AP*GP)-3' | | Authors: | Nielsen, J.T, Stein, P.C, Petersen, M. | | Deposit date: | 2003-07-23 | | Release date: | 2003-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of an Alpha-L-Lna:RNA Hybrid: Structural Implications for Rnase H Recognition
Nucleic Acids Res., 31, 2003
|
|
2YCW
 
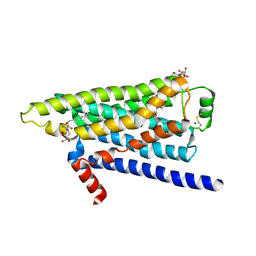 | | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND ANTAGONIST CARAZOLOL | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, BETA-1 ADRENERGIC RECEPTOR, HEGA-10, ... | | Authors: | Moukhametzianov, R, Warne, T, Edwards, P.C, Serrano-Vega, M.J, Leslie, A.G.W, Tate, C.G, Schertler, G.F.X. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two Distinct Conformations of Helix 6 Observed in Antagonist-Bound Structures of a Beta-1- Adrenergic Receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1P7Z
 
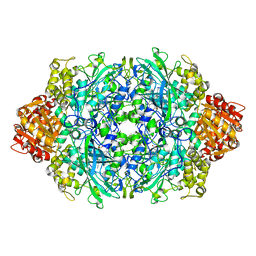 | | Crystal structure of the D181S variant of catalase HPII from E. coli | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
1P7Y
 
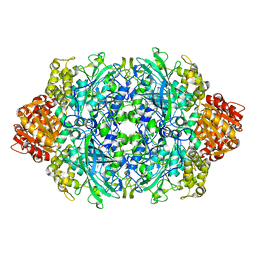 | | Crystal structure of the D181A variant of catalase HPII from E. coli | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
1SIY
 
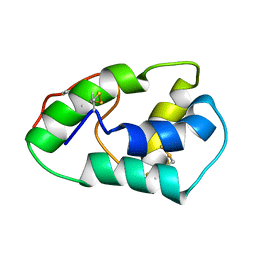 | | NMR structure of mung bean non-specific lipid transfer protein 1 | | Descriptor: | Nonspecific lipid-transfer protein 1 | | Authors: | Lin, K.F, Liu, Y.N, Hsu, S.T.D, Samuel, D, Cheng, C.S, Bonvin, A.M.J.J, Lyu, P.C. | | Deposit date: | 2004-03-02 | | Release date: | 2005-04-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Characterization and Structural Analyses of Nonspecific Lipid Transfer Protein 1 from Mung Bean
Biochemistry, 44, 2005
|
|
1P80
 
 | | Crystal structure of the D181Q variant of catalase HPII from E. coli | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
1RSZ
 
 | | Structure of human purine nucleoside phosphorylase in complex with DADMe-Immucillin-H and sulfate | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Shi, W, Lewandowicz, A, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-12-10 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural comparison of human and malarial purine nucleoside phosphorylases
To be Published
|
|
1NG4
 
 | | Structure of ThiO (glycine oxidase) from Bacillus subtilis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glycine oxidase, HYDROGEN PEROXIDE, ... | | Authors: | Settembre, E.C, Dorrestein, P.C, Park, J, Augustine, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Studies on ThiO, a Glycine Oxidase Essential for Thiamin Biosynthesis in Bacillus subtilis
Biochemistry, 42, 2003
|
|
1NG3
 
 | | Complex of ThiO (glycine oxidase) with acetyl-glycine | | Descriptor: | ACETYLAMINO-ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Glycine oxidase, ... | | Authors: | Settembre, E.C, Dorrestein, P.C, Park, J, Augustine, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Studies on ThiO, a Glycine Oxidase Essential for Thiamin Biosynthesis in Bacillus subtilis
Biochemistry, 42, 2003
|
|
1B1C
 
 | | CRYSTAL STRUCTURE OF THE FMN-BINDING DOMAIN OF HUMAN CYTOCHROME P450 REDUCTASE AT 1.93A RESOLUTION | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, PROTEIN (NADPH-CYTOCHROME P450 REDUCTASE) | | Authors: | Zhao, Q, Modi, S, Smith, G, Paine, M, Mcdonagh, P.D, Wolf, C.R, Tew, D, Lian, L.-Y, Roberts, G.C.K, Driessen, H.P.C. | | Deposit date: | 1998-11-19 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 A resolution.
Protein Sci., 8, 1999
|
|
