5BXA
 
 | | Structure of PslG from Pseudomonas aeruginosa in complex with mannose | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Baker, P, Little, D.J, Howell, P.L. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of the Pseudomonas aeruginosa Glycoside Hydrolase PslG Reveals That Its Levels Are Critical for Psl Polysaccharide Biosynthesis and Biofilm Formation.
J.Biol.Chem., 290, 2015
|
|
6Z9K
 
 | | CAP domain of Enterococcal PrgA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PrgA | | Authors: | Berntsson, R.P.A, Schmitt, A. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enterococcal PrgA Extends Far Outside the Cell and Provides Surface Exclusion to Protect against Unwanted Conjugation.
J.Mol.Biol., 432, 2020
|
|
6NJ6
 
 | |
7RAA
 
 | | Designed StabIL-2 seq15 | | Descriptor: | Interleukin-2, MAGNESIUM ION | | Authors: | Jude, K.M, Chu, A.E, Huang, P.-S, Garcia, K.C. | | Deposit date: | 2021-06-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Interleukin-2 superkines by computational design.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
1LHD
 
 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BOROLYS-OH | | Descriptor: | AC-(D)PHE-PRO-BOROLYS-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
6NLZ
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment degradation product B9D | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, [(1R,2S)-2-(2-hydroxybenzene-1-carbonyl)cyclopentyl]acetic acid, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment degradation product B9D
To Be Published
|
|
1ERI
 
 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | Authors: | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | Deposit date: | 1994-05-18 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
6NN0
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with 2'-deoxycytidine and fragment degradation product B9D | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with 2'-deoxycytidine and B9D
To Be Published
|
|
6JJQ
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.99 A resolution. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Bairagya, H.R, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-02-26 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.99 A resolution.
To Be Published
|
|
1E6S
 
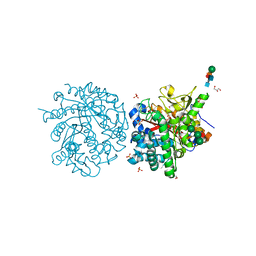 | | MYROSINASE FROM SINAPIS ALBA with bound gluco-hydroximolactam and sulfate | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burmeister, W.P. | | Deposit date: | 2000-08-23 | | Release date: | 2000-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution X-Ray Crystallography Shows that Ascorbate is a Cofactor for Myrosinase and Substitutes for the Function of the Catalytic Base
J.Biol.Chem., 275, 2000
|
|
1O4O
 
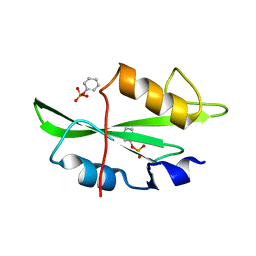 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PHENYLPHOSPHATE. | | Descriptor: | PHENYL DIHYDROGEN PHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
7RI1
 
 | |
6ZDT
 
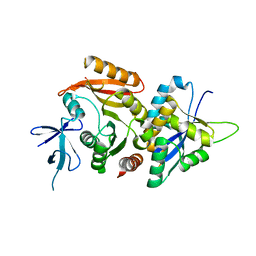 | | Crystal structure of eukaryotic Fibrillarin with Nop56 N-terminal domain | | Descriptor: | Nucleolar protein 56, rRNA 2'-O-methyltransferase fibrillarin | | Authors: | Hoefler, S, Lukat, P, Carlomagno, T, Blankenfeldt, W. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | High-resolution structure of eukaryotic Fibrillarin interacting with Nop56 amino-terminal domain.
Rna, 27, 2021
|
|
4ODH
 
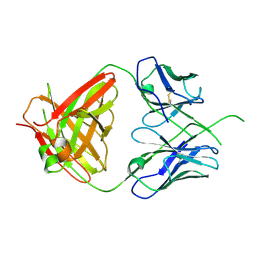 | | Crystal structure of human Fab CAP256-VRC26.UCA, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.UCA heavy chain, CAP256-VRC26.UCA light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-10 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4OVB
 
 | | Crystal structure of Oncogenic Suppression Activity Protein - A Plasmid Fertility Inhibition Factor, Gold (I) Cyanide derivative | | Descriptor: | GLYCEROL, GOLD (I) CYANIDE ION, PHOSPHATE ION, ... | | Authors: | Maindola, P, Goyal, P, Arulandu, A. | | Deposit date: | 2014-02-21 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids
Nat Commun, 5, 2014
|
|
5NIF
 
 | | Yeast 20S proteasome in complex with Blm-pep activator | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Witkowska, J, Grudnik, P, Golik, P, Dubin, G, Jankowska, E. | | Deposit date: | 2017-03-23 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a low molecular weight activator Blm-pep with yeast 20S proteasome - insights into the enzyme activation mechanism.
Sci Rep, 7, 2017
|
|
6BLH
 
 | | RSV G central conserved region bound to Fab CB017.5 | | Descriptor: | 1,2-ETHANEDIOL, Fab CB017.5 heavy chain, Fab CB017.5 light chain, ... | | Authors: | Jones, H.G, McLellan, J.S, Langedijk, J.P. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition of the central conserved region of RSV G by neutralizing human antibodies.
PLoS Pathog., 14, 2018
|
|
6RJL
 
 | | Fragment AZ-018 binding at the TAZpS89/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-(3-azanylpropyl)-4-phenyl-thiophene-2-carboximidamide, TAZpS89 | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-27 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
8QMX
 
 | | OPR3 wildtype in complex with NADPH4 | | Descriptor: | 12-oxophytodienoate reductase 3, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Loop 6 and the beta-hairpin flap are structural hotspots that determine cofactor specificity in the FMN-dependent family of ene-reductases.
Febs J., 291, 2024
|
|
5FD9
 
 | | X-ray Crystal Structure of ESCRT-III Snf7 core domain (conformation B) | | Descriptor: | Vacuolar-sorting protein SNF7 | | Authors: | Tang, S, Henne, W.M, Borbat, P.P, Buchkovich, N.J, Freed, J.H, Mao, Y, Fromme, J.C, Emr, S.D. | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments.
Elife, 4, 2015
|
|
2WD2
 
 | | A chimeric microtubule disruptor with efficacy on a taxane resistant cell line | | Descriptor: | 7-methoxy-2-(3-methoxybenzyl)-1,2,3,4-tetrahydroisoquinolin-6-yl sulfamate, CARBONIC ANHYDRASE 2, FORMIC ACID, ... | | Authors: | Leese, M.P, Jourdan, F.L, Kimberley, M.R, Cozier, G.E, Regis-Lydi, S, Foster, P.A, Newman, S.P, Thiyagarajan, N, Acharya, K.R, Ferrandis, E, Purohit, A, Reed, M.J, Potter, B.V.L. | | Deposit date: | 2009-03-19 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Chimeric Microtubule Disruptors.
Chem.Commun.(Camb.), 46, 2010
|
|
6MNZ
 
 | | Crystal structure of RibBX, a two domain 3,4-dihydroxy-2-butanone 4-phosphate synthase from A. baumannii. | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Wang, J, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Multi-metal Restriction by Calprotectin Impacts De Novo Flavin Biosynthesis in Acinetobacter baumannii.
Cell Chem Biol, 26, 2019
|
|
7U0B
 
 | | Crystal structure of broadly neutralizing antibody HEPC3.1 | | Descriptor: | HEPC3.1 Fab Heavy Chain, HEPC3.1 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Computational identification of HCV neutralizing antibodies with a common HCDR3 disulfide bond motif in the antibody repertoires of infected individuals.
Nat Commun, 13, 2022
|
|
1PLR
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
