8PJL
 
 | |
6X2B
 
 | |
7RTI
 
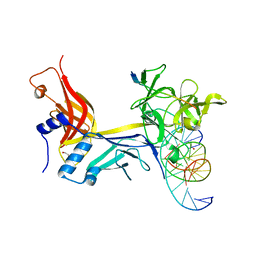 | | X-ray structure of RBPJ-L3MBTL3(dT62)-DNA complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*CP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Hall, D.P, Kovall, R.A, Yuan, Z. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure, binding and function of a Notch transcription complex involving RBPJ and the epigenetic reader protein L3MBTL3.
Nucleic Acids Res., 50, 2022
|
|
8B45
 
 | | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4 | | Descriptor: | 1,2-ETHANEDIOL, CC-Tri-(UbUc)4, SODIUM ION, ... | | Authors: | Kumar, P, Martin, F.J.O, Dawson, W.M, Zieleniewski, F, Woolfson, D.N. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4
To Be Published
|
|
6KVC
 
 | | MoeE5 in complex with UDP-glucose and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
8BWG
 
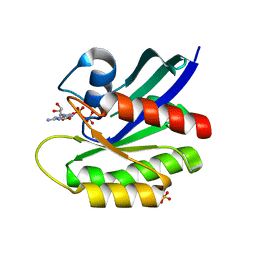 | | HRas (1-166) Y64 phosphorylation | | Descriptor: | GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Baumann, P, Jin, Y. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Far-reaching effects of tyrosine64 phosphorylation on Ras revealed with BeF 3 - complexes.
Commun Chem, 7, 2024
|
|
1MSJ
 
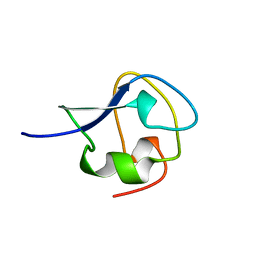 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15V | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
6WT4
 
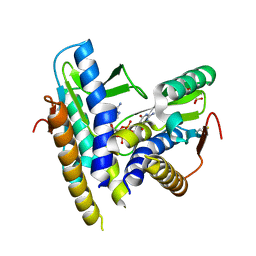 | | Structure of a bacterial STING receptor from Flavobacteriaceae sp. in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Bacterial STING, SULFATE ION | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
6HWG
 
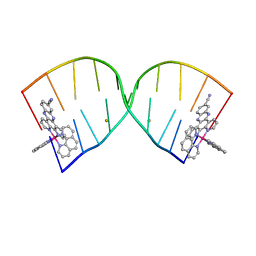 | |
6WP7
 
 | |
7R6A
 
 | |
7R69
 
 | |
6L1S
 
 | | Crystal structure of DUSP22 mutant_C88S | | Descriptor: | Dual specificity protein phosphatase 22, PHOSPHATE ION | | Authors: | Lai, C.H, Chang, C.C, Lyu, P.C. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3611 Å) | | Cite: | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
3V5A
 
 | | Crystal Structure of C-lobe of Bovine Lactoferrin Complexed with Gamma Amino Butyric Acid at 1.44 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-TERMINAL PEPTIDE OF LACTOTRANSFERRIN, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-12-16 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal Structure of C-lobe of Bovine Lactoferrin Complexed with Gamma Amino Butyric Acid at 1.44 A Resolution
To be Published
|
|
2J8M
 
 | | Structure of P. aeruginosa acetyltransferase PA4866 | | Descriptor: | ACETYLTRANSFERASE PA4866 FROM P. AERUGINOSA, AZIDE ION, GLYCEROL, ... | | Authors: | Davies, A.M, Tata, R, Beavil, R.L, Sutton, B.J, Brown, P.R. | | Deposit date: | 2006-10-26 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | L-Methionine Sulfoximine, But not Phosphinothricin, is a Substrate for an Acetyltransferase (Gene Pa4866) from Pseudomonas Aeruginosa: Structural and Functional Studies.
Biochemistry, 46, 2007
|
|
6X2C
 
 | |
7R0J
 
 | | Structure of the V2 receptor Cter-arrestin2-ScFv30 complex | | Descriptor: | Arrestin2, ScFv30, V2R Cter | | Authors: | Bous, J, Fouillen, A, Trapani, S, Granier, S, Mouillac, B, Bron, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex.
Sci Adv, 8, 2022
|
|
6LAN
 
 | | Structure of CCDC50 and LC3B complex | | Descriptor: | Coiled-coil domain-containing protein 50,Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Liu, L, Li, J, Hou, P. | | Deposit date: | 2019-11-12 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection.
Cell Res., 31, 2021
|
|
6RCA
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with 2.2'-anhydrouridine at 1.34 A | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-Anhydro-(1-beta-D-ribofuranosyl)uracil, CHLORIDE ION, ... | | Authors: | Prokofev, I.I, Eistrikh-Geller, P.A, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Lashkov, A.A. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | X-Ray Structure and Molecular Dynamics Study of Uridine Phosphorylase from Vibrio cholerae in Complex with 2,2'-Anhydrouridine
Crystallography Reports, 2020
|
|
7R0C
 
 | | Structure of the AVP-V2R-arrestin2-ScFv30 complex | | Descriptor: | AVP, Arrestin2, ScFv30, ... | | Authors: | Bous, J, Fouillen, A, Trapani, S, Granier, S, Mouillac, B, Bron, P. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex.
Sci Adv, 8, 2022
|
|
8JUA
 
 | | Multifunctional cytochrome P450 enzyme IkaD from Streptomyces sp. ZJ306, in complex with epoxyikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,13R,15S,16R,17S,19Z,26S)-11-ethyl-2-hydroxy-10-methyl-22,27-diaza-14 oxahexacyclo[24.2.1.05,17.07,16.013,15.08,12]nonacosa-1(2),3,19-triene-21,28,29-trione, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.00001121 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BBC
 
 | |
8BBA
 
 | |
8BB9
 
 | |
6DWI
 
 | |
