5RF7
 
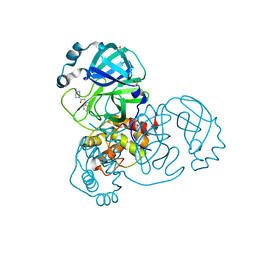 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z316425948_minor | | Descriptor: | 1-(4-methylpiperazin-1-yl)-2-(1H-pyrrolo[2,3-b]pyridin-3-yl)ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFM
 
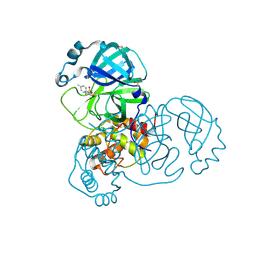 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102539 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(3R)-1,1-dioxo-2,3-dihydro-1H-1lambda~6~-thiophen-3-yl]-N-(4-methylphenyl)acetamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RG0
 
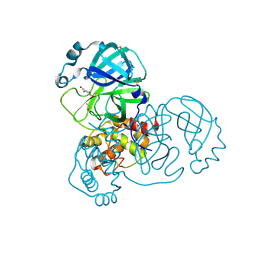 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102535 | | Descriptor: | 1,1'-(piperazine-1,4-diyl)di(ethan-1-one), 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RGG
 
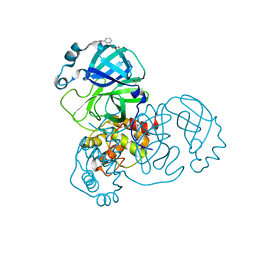 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2856434890 (Mpro-x0165) | | Descriptor: | 3C-like proteinase, 4-methyl-N-phenylpiperazine-1-carboxamide, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RGL
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102962 (Mpro-x0705) | | Descriptor: | 1-[4-(4-methylbenzene-1-carbonyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RHC
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Cov_HetLib053 (Mpro-x2119) | | Descriptor: | (E)-1-(1H-imidazol-2-yl)methanimine, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, Keeley, A, Keseru, G.M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-16 | | Release date: | 2020-06-10 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
8TGB
 
 | | Crystal structure of root lateral formation protein (RLF) b5-domain from Oryza sativa | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, root lateral formation protein (RLF) | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Benson, D.R. | | Deposit date: | 2023-07-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The N-terminal intrinsically disordered region of Ncb5or docks with the cytochrome b 5 core to form a helical motif that is of ancient origin.
Proteins, 92, 2024
|
|
8TJJ
 
 | | SAM-dependent methyltransferase RedM bound to SAM | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, RedM, ... | | Authors: | Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2023-07-22 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of methyltransferase RedM that forms the dimethylpyrrolinium of the bisindole reductasporine.
J.Biol.Chem., 300, 2023
|
|
6TWI
 
 | | Crystal structure of the haemagglutinin mutant (Gln226Leu, Gly228Ser) from an H10N7 seal influenza virus isolated in Germany in complex with avian receptor analogue 3'-SLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-13 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
1A5O
 
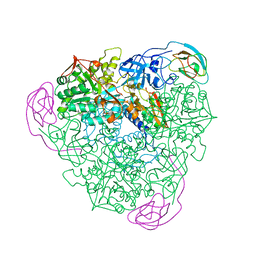 | | K217C VARIANT OF KLEBSIELLA AEROGENES UREASE, CHEMICALLY RESCUED BY FORMATE AND NICKEL | | Descriptor: | FORMIC ACID, NICKEL (II) ION, UREASE (ALPHA SUBUNIT), ... | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1QIQ
 
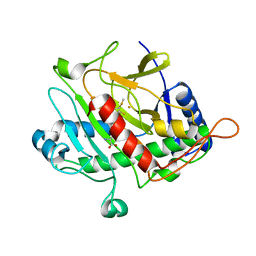 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACmC Fe COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, N-[N-[2-AMINO-6-OXO-HEXANOIC ACID-6-YL]CYSTEINYL]-S-METHYLCYSTEINE, ... | | Authors: | Rutledge, P.J, Clifton, I.J, Burzlaff, N.I, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction.
Nature, 401, 1999
|
|
1A5M
 
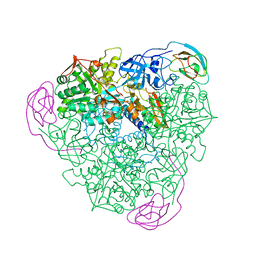 | | K217A VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
6OSM
 
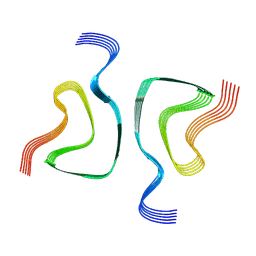 | |
6I99
 
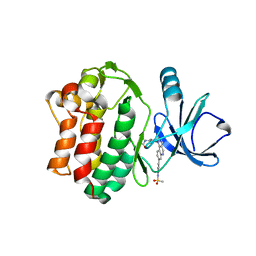 | | Bone Marrow Tyrosine Kinase in Chromosome X in complex with a newly designed covalent inhibitor JS24 | | Descriptor: | Cytoplasmic tyrosine-protein kinase BMX, ~{N}-[2-methyl-5-[8-[4-(methylsulfonylamino)phenyl]-2-oxidanylidene-benzo[h][1,6]naphthyridin-1-yl]phenyl]-3-oxidanyl-propanamide | | Authors: | Sousa, B.B, Matias, P.M, Marques, M.C, Seixas, J.D, Bernardes, G.J.L. | | Deposit date: | 2018-11-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural and biophysical insights into the mode of covalent binding of rationally designed potent BMX inhibitors
Chem.Biol., 2020
|
|
4ZPW
 
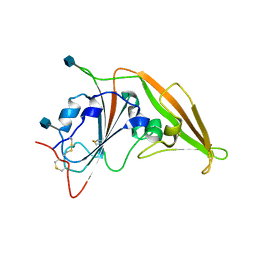 | | Structure of unbound MERS-CoV spike receptor-binding domain (England1 strain). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, Spike glycoprotein | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
4WSX
 
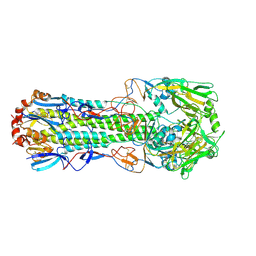 | | The crystal structure of hemagglutinin from A/Jiangxi-Donghu/346/2013 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-10-28 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and receptor binding preferences of recombinant hemagglutinins from avian and human h6 and h10 influenza a virus subtypes.
J.Virol., 89, 2015
|
|
2MCM
 
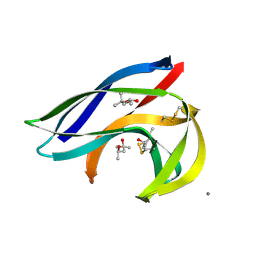 | | MACROMOMYCIN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Van Roey, P. | | Deposit date: | 1991-05-08 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure analysis of auromomycin apoprotein (macromomycin) shows importance of protein side chains to chromophore binding selectivity.
Proc.Natl.Acad.Sci.Usa, 86, 1989
|
|
7A6X
 
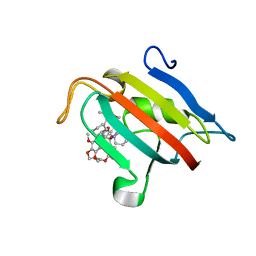 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 56 | | Descriptor: | (2S,9S,12R)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-24,27-dimethoxy-11,18,22-trioxa-4-azatetracyclo[21.2.2.113,17.04,9]octacosa-1(25),13(28),14,16,23,26-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6U1J
 
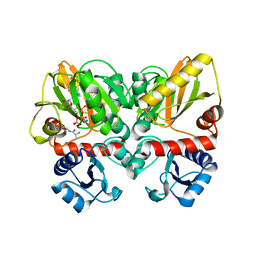 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ADP, phosphate, D-ala-D-ala, Mg2+ and K+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B, Thompson, A.P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
5R80
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z18197050 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, methyl 4-sulfamoylbenzoate | | Authors: | Fearon, D, Powell, A.J, Douangamath, A, Owen, C.D, Wild, C, Krojer, T, Lukacik, P, Strain-Damerell, C.M, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
6UH8
 
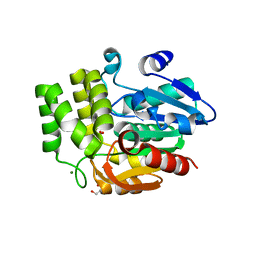 | | Crystal structure of DAD2 N242I mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Decreased Apical Dominance 2, GLYCEROL, ... | | Authors: | Sharma, P, Hamiaux, C, Snowden, K.C. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners.
J.Biol.Chem., 295, 2020
|
|
7A6W
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 33-(Z) | | Descriptor: | (2S,9S,12R,20Z)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-28,31-dimethoxy-11,18,23,26-tetraoxa-4-azatetracyclo[25.2.2.113,17.04,9]dotriaconta-1(29),13(32),14,16,20,27,30-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
5HUG
 
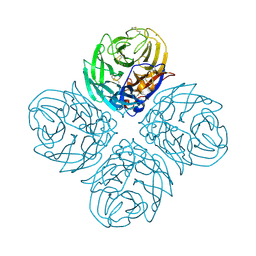 | | The crystal structure of neuraminidase from A/American green-winged teal/Washington/195750/2014 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, H, Carney, P.J, Guo, Z, Chang, J.C, Stevens, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Characterizations of Surface Proteins Hemagglutinin and Neuraminidase from Recent H5Nx Avian Influenza Viruses.
J.Virol., 90, 2016
|
|
3HWN
 
 | | CATHEPSIN L with AZ13010160 | | Descriptor: | Cathepsin L1, Nalpha-[(3-tert-butyl-1-methyl-1H-pyrazol-5-yl)carbonyl]-N-[(2E)-2-iminoethyl]-3-{5-[(Z)-iminomethyl]-1,3,4-oxadiazol-2-yl}-L-phenylalaninamide | | Authors: | Kenny, P, Morley, A. | | Deposit date: | 2009-06-18 | | Release date: | 2009-09-15 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Design of selective Cathepsin inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2MLL
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-16 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
