5MFO
 
 | | Designed armadillo repeat protein YIIIM3AIII | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, YIIIM3AIII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
6H9I
 
 | | Csf5, CRISPR-Cas type IV Cas6 crRNA endonuclease | | Descriptor: | Csf5, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-08-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum.
Nat Microbiol, 4, 2019
|
|
7D52
 
 | | Crystal structure of yak lactoperoxidase with a disordered propionic group of heme moiety at 2.20 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, P.K, Rani, C, Ahmad, N, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-09-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
7OYE
 
 | |
7OZI
 
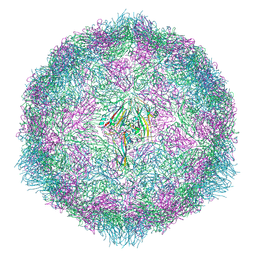 | |
6HDF
 
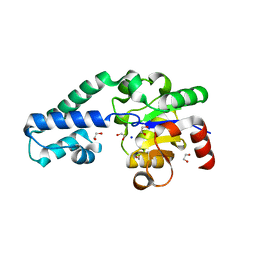 | | D170N variant of beta-phosphoglucomutase from Lactococcus lactis in an open conformer to 1.4 A. | | Descriptor: | 1,2-ETHANEDIOL, Beta-phosphoglucomutase, SODIUM ION | | Authors: | Wood, H.P, Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
7WYJ
 
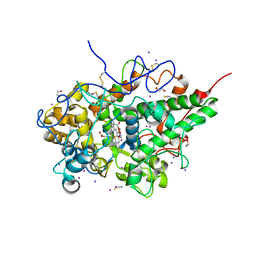 | | Structure of the complex of lactoperoxidase with nitric oxide catalytic product nitrite at 1.89 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Pandey, N, Singh, A.K, Sinha, M, Singh, R.P, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural evidence of the conversion of nitric oxide (NO) to nitrite ion (NO2-) by lactoperoxidase (LPO): Structure of the complex of LPO with NO2- at 1.89 angstrom resolution
J.Inorg.Biochem., 247, 2023
|
|
7P2X
 
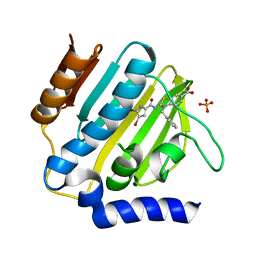 | | E.coli GyrB24 with inhibitor KOB20 (EBL2583) | | Descriptor: | (2Z)-2-[[4,5-bis(bromanyl)-1H-pyrrol-2-yl]carbonylimino]-3-(phenylmethyl)-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Benek, O, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | E.coli GyrB24 with inhibitor KOB20 (EBL2583)
TO BE PUBLISHED
|
|
9B76
 
 | |
9B75
 
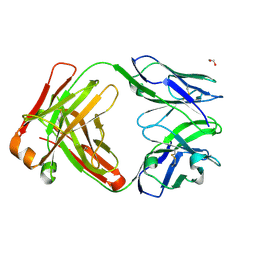 | |
8SZQ
 
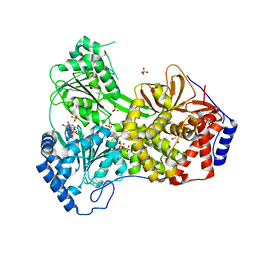 | | Cat DHX9 bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7DH7
 
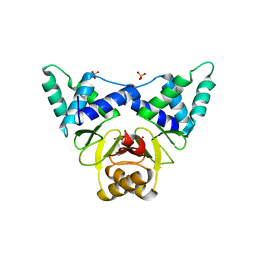 | | Crystal structure of apo XcZur | | Descriptor: | PHOSPHATE ION, Transcriptional regulator fur family, ZINC ION | | Authors: | Liu, F.M, Su, Z.H, Chen, P, Tian, X.L, Wu, L.J, Tang, D.J, Li, P.F, Deng, H.T, Tang, J.L, Ming, Z.H. | | Deposit date: | 2020-11-13 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for zinc-induced activation of a zinc uptake transcriptional regulator.
Nucleic Acids Res., 49, 2021
|
|
4YDI
 
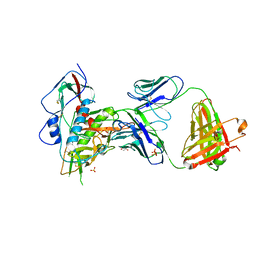 | | Crystal structure of broad and potently neutralizing VRC01-class antibody Z258-VRC27.01, isolated from human donor Z258, in complex with HIV-1 gp120 from clade A strain Q23.17 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.452 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
7OZK
 
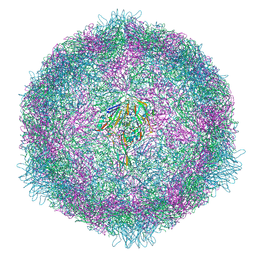 | | CryoEM structure of human enterovirus 70 in complex with Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Fuzik, T, Plevka, P, Moravcova, J. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Structure of Human Enterovirus 70 and Its Inhibition by Capsid-Binding Compounds.
J.Virol., 96, 2022
|
|
7OZL
 
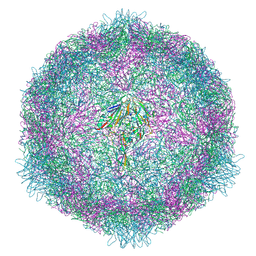 | | CryoEM structure of human enterovirus 70 in complex with WIN51711 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Fuzik, T, Plevka, P, Moravcova, J. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structure of Human Enterovirus 70 and Its Inhibition by Capsid-Binding Compounds.
J.Virol., 96, 2022
|
|
8TO9
 
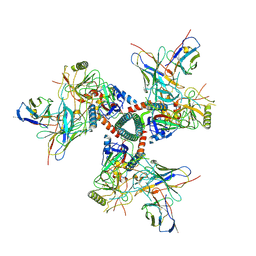 | | Cryo-EM structure of TRNM-f*01 Fab in complex with HIV-1 Env trimer ConC SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Roark, R.S, Morano, N.C, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Broad and potent HIV-1 neutralization in fusion peptide-primed SHIV-boosted macaques
To Be Published
|
|
8VON
 
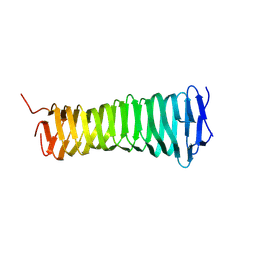 | |
8GIH
 
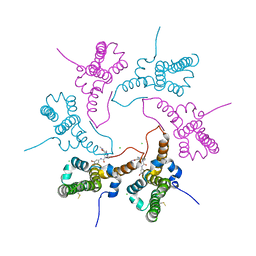 | | Structure of Hepatitis B Virus Capsid Y132A mutant in complex with Compound 24 | | Descriptor: | (6S,8R)-N-(3-bromo-4-fluorophenyl)-8-fluoro-10-methyl-11-oxo-1,3,4,7,8,9,10,11-octahydro-2H-pyrido[4',3':3,4]pyrazolo[1,5-a][1,4]diazepine-2-carboxamide, CHLORIDE ION, Capsid protein | | Authors: | Shaffer, P.L. | | Deposit date: | 2023-03-14 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Diazepinone HBV capsid assembly modulators.
Bioorg Med Chem Lett, 72, 2022
|
|
6XUB
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with monovinyl monopropionyl deuteroheme | | Descriptor: | Chlorite dismutase, harderoheme (III) | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
7UJX
 
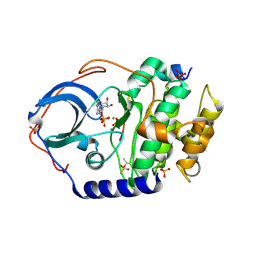 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 2.4 Angstrom data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wych, D.C, Aoto, P.C, Wall, M.E. | | Deposit date: | 2022-03-31 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5DO5
 
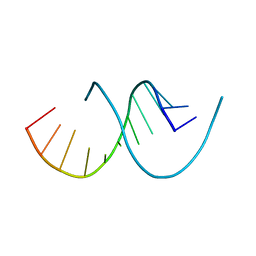 | |
6XWU
 
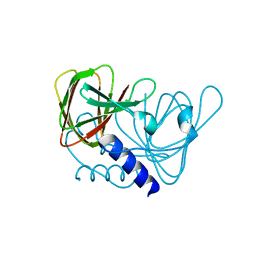 | | Crystal structure of drosophila melanogaster CENP-C cumin domain | | Descriptor: | RE68959p | | Authors: | Jeyaprakash, A.A, Medina-Pritchard, B, Lazou, V, Zou, J, Byron, O, Abad, M.A, Rappsilber, J, Heun, P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1.
Embo J., 39, 2020
|
|
8OHD
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ8
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
7V0G
 
 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 1.63 Angstrom data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wych, D.C, Aoto, P.C, Wall, M.E. | | Deposit date: | 2022-05-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
