4ZV6
 
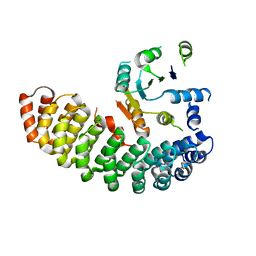 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | Descriptor: | AlphaRep-7, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
6U69
 
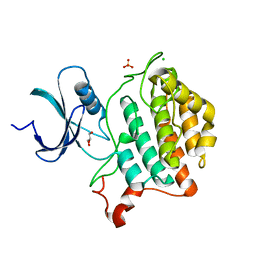 | | Crystal structure of Yck2 from Candida albicans, apoenzyme | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Overcoming Fungal Echinocandin Resistance through Inhibition of the Non-essential Stress Kinase Yck2.
Cell Chem Biol, 27, 2020
|
|
150D
 
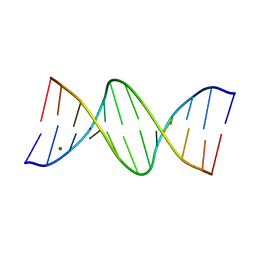 | | GUANINE.1,N6-ETHENOADENINE BASE-PAIRS IN THE CRYSTAL STRUCTURE OF D(CGCGAATT(EDA)GCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(EDA)P*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, McAuley-Hecht, K.E, Gibson, N.J, Brown, T, Watson, W.P, Hunter, W.N. | | Deposit date: | 1993-12-02 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Guanine-1,N6-ethenoadenine base pairs in the crystal structure of d(CGCGAATT(epsilon dA)GCG).
Biochemistry, 33, 1994
|
|
165D
 
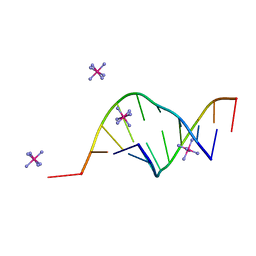 | | THE STRUCTURE OF A MISPAIRED RNA DOUBLE HELIX AT 1.6 ANGSTROMS RESOLUTION AND IMPLICATIONS FOR THE PREDICTION OF RNA SECONDARY STRUCTURE | | Descriptor: | DNA/RNA (5'-R(*GP*CP*UP*UP*CP*GP*GP*CP*)-D(*(BRU))-3'), RHODIUM HEXAMINE ION | | Authors: | Cruse, W, Saludjian, P, Biala, E, Strazewski, P, Prange, T, Kennard, O. | | Deposit date: | 1994-03-21 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a mispaired RNA double helix at 1.6-A resolution and implications for the prediction of RNA secondary structure.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
4ZWY
 
 | | Human Carbonic Anhydrase II in complex with a glucosyl sulfamate inhibitor | | Descriptor: | (6S)-1,3,4,5-tetra-O-acetyl-2,6-anhydro-6-{[5-(sulfamoyloxy)pentyl]sulfamoyl}-L-altritol, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Mahon, B.P, Lomelino, C.L, Driscoll, J.M, McKenna, R. | | Deposit date: | 2015-05-19 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping Selective Inhibition of the Cancer-Related Carbonic Anhydrase IX Using Structure-Activity Relationships of Glucosyl-Based Sulfamates.
J.Med.Chem., 58, 2015
|
|
4ZZX
 
 | | Structure of PARP2 catalytic domain bound to an isoindolinone inhibitor | | Descriptor: | 2-(3-methoxypropyl)-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, POLY [ADP-RIBOSE] POLYMERASE 2 | | Authors: | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
4ZWC
 
 | | Crystal structure of maltose-bound human GLUT3 in the outward-open conformation at 2.6 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
4ZX1
 
 | | Engineered Carbonic Anhydrase IX mimic in complex with a glucosyl sulfamate inhibitor | | Descriptor: | (6R)-5-O-acetyl-2,6-anhydro-6-{[4-(sulfamoyloxy)piperidin-1-yl]sulfonyl}-L-glucitol, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Mahon, B.P, Lomelino, C.L, Salguero, A.L, McKenna, R. | | Deposit date: | 2015-05-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Mapping Selective Inhibition of the Cancer-Related Carbonic Anhydrase IX using Structure-Activity Relationships of Glucosyl-Based Sulfamates
J. Med. Chem., 58, 2015
|
|
4ZW9
 
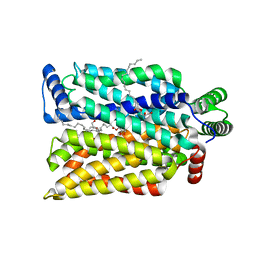 | | Crystal structure of human GLUT3 bound to D-glucose in the outward-occluded conformation at 1.5 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
4ZXZ
 
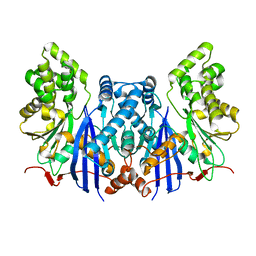 | |
5A2B
 
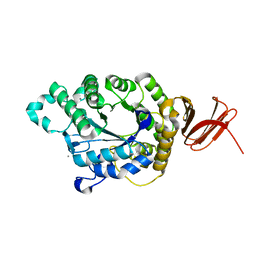 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ANOXYBACILLUS ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
4ZTP
 
 | |
4ZX0
 
 | | Human Carbonic Anhydrase II in complex with a glucosyl sulfamate inhibitor | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION, ... | | Authors: | Mahon, B.P, Lomelino, C.L, Pinard, M.A, McKenna, R. | | Deposit date: | 2015-05-19 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping Selective Inhibition of the Cancer-Related Carbonic Anhydrase IX using Structure-Activity Relationships of Glucosyl-Based Sulfamates
J. Med. Chem., 58, 2015
|
|
5A6E
 
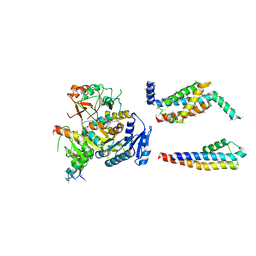 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, ... | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
4ZZN
 
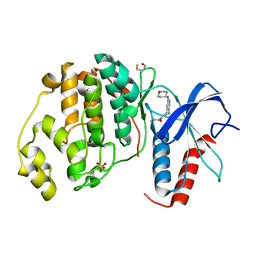 | | Human ERK2 in complex with an inhibitor | | Descriptor: | 2-[[5-chloranyl-2-(oxan-4-ylamino)pyridin-4-yl]amino]-N-methyl-benzamide, MITOGEN-ACTIVATED PROTEIN KINASE 1, SULFATE ION | | Authors: | Ward, R.A, Colclough, N, Challinor, M, Debreczeni, J.E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, James, M, Jones, C.D, Jones, C.R, Renshaw, J, Roberts, K, Snow, L, Tonge, M, Yeung, K. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-27 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of Erk1/2.
J.Med.Chem., 58, 2015
|
|
5A92
 
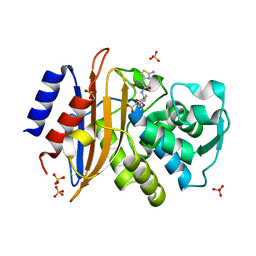 | | 15K X-ray structure with Cefotaxime: Exploring the Mechanism of beta- Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | Descriptor: | BETA-LACTAMASE CTX-M-97, CEFOTAXIME, C3' cleaved, ... | | Authors: | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-16 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
5AFG
 
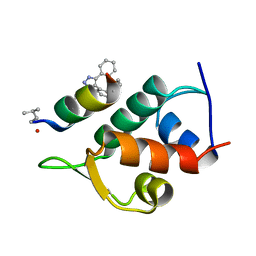 | | Structure of the Stapled Peptide Bound to Mdm2 | | Descriptor: | 1,8-DIETHYL-1,8-DIHYDRODIBENZO[3,4:7,8][1,2,3]TRIAZOLO[4',5':5,6]CYCLOOCTA[1,2-D][1,2,3]TRIAZOLE, E3 UBIQUITIN-PROTEIN LIGASE MDM2, STAPLED PEPTIDE | | Authors: | Lau, Y.H, Wu, Y, Rossmann, M, de Andrade, P, Tan, Y.S, McKenzie, G.J, Venkitaraman, A.R, Hyvonen, M, Spring, D.R. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Double Strain-Promoted Macrocyclization for the Rapid Selection of Cell-Active Stapled Peptides.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AF6
 
 | | Structure of Lys33-linked diUb bound to Trabid NZF1 | | Descriptor: | TRABID, UBIQUITIN, ZINC ION | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
1A5M
 
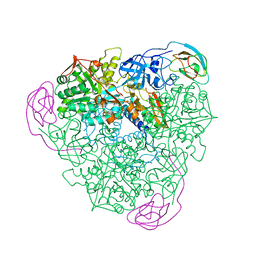 | | K217A VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5O
 
 | | K217C VARIANT OF KLEBSIELLA AEROGENES UREASE, CHEMICALLY RESCUED BY FORMATE AND NICKEL | | Descriptor: | FORMIC ACID, NICKEL (II) ION, UREASE (ALPHA SUBUNIT), ... | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5N
 
 | | K217A VARIANT OF KLEBSIELLA AEROGENES UREASE, CHEMICALLY RESCUED BY FORMATE AND NICKEL | | Descriptor: | FORMIC ACID, NICKEL (II) ION, UREASE (ALPHA SUBUNIT), ... | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5K
 
 | | K217E VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1A5L
 
 | | K217C VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
1AIE
 
 | |
4UER
 
 | | 40S-eIF1-eIF1A-eIF3-eIF3j translation initiation complex from Lachancea kluyveri | | Descriptor: | 18S RRNA, EIF1, EIF1A, ... | | Authors: | Aylett, C.H.S, Boehringer, D, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-12-18 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | Structure of a Yeast 40S-Eif1-Eif1A-Eif3-Eif3J Initiation Complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
