5REZ
 
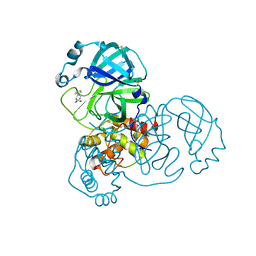 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with POB0129 | | Descriptor: | (1R,2S)-2-(thiophen-3-yl)cyclopentane-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFE
 
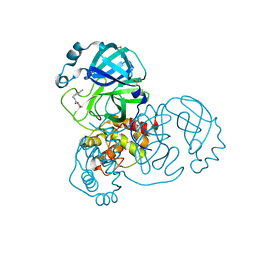 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z509756472 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(4-cyanophenyl)methyl]morpholine-4-carboxamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
224D
 
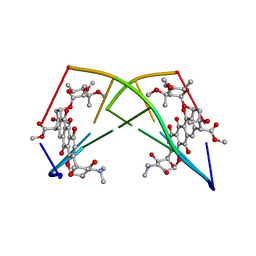 | | DNA-DRUG REFINEMENT: A COMPARISON OF THE PROGRAMS NUCLSQ, PROLSQ, SHELXL93 AND X-PLOR, USING THE LOW TEMPERATURE D(TGATCA)-NOGALAMYCIN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Schuerman, G.S, Smith, C.K, Turkenburg, J.P, Dettmar, A.N, Van Meervelt, L, Moore, M.H. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DNA-drug refinement: a comparison of the programs NUCLSQ, PROLSQ, SHELXL93 and X-PLOR, using the low-temperature d(TGATCA)-nogalamycin structure.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2Y26
 
 | | Transmission defective mutant of Grapevine Fanleaf virus | | Descriptor: | COAT PROTEIN | | Authors: | Schellenberger, P, Sauter, C, Lorber, B, Bron, P, Trapani, S, Bergdoll, M, Marmonier, A, Schmitt-Keichinger, C, Lemaire, O, Demangeat, G, Ritzenthaler, C. | | Deposit date: | 2010-12-13 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into Viral Determinants of Nematode Mediated Grapevine Fanleaf Virus Transmission.
Plos Pathog., 7, 2011
|
|
1XZV
 
 | |
4JV0
 
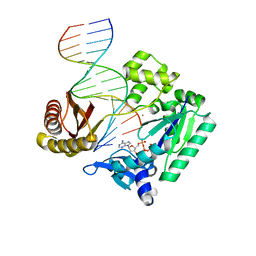 | | Ring-Opening of the -OH-PdG Adduct in Ternary Complexes with the Sulfolobus solfataricus DNA polymerase Dpo4 | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), ... | | Authors: | Banerjee, S, Shanmugam, G, Stone, M.P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Ring-Opening of the gamma-OH-PdG Adduct Promotes Error-Free Bypass by the Sulfolobus solfataricus DNA Polymerase Dpo4.
Chem.Res.Toxicol., 26, 2013
|
|
1XID
 
 | |
1XIF
 
 | |
2LH2
 
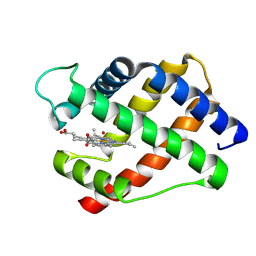 | | X-RAY STRUCTURAL INVESTIGATION OF LEGHEMOGLOBIN. VI. STRUCTURE OF ACETATE-FERRILEGHEMOGLOBIN AT A RESOLUTION OF 2.0 ANGSTROMS (RUSSIAN) | | Descriptor: | LEGHEMOGLOBIN (AQUO MET), OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Harutyunyan, E.H, Kuranova, I.P, Borisov, V.V, Sosfenov, N.I, Pavlovsky, A.G, Grebenko, A.I, Konareva, N.V. | | Deposit date: | 1982-04-23 | | Release date: | 1983-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structural Investigation of Leghemoglobin. Vi. Structure of Acetate-Ferrileghemoglobin at a Resolution of 2.0 Angstroms (Russian)
Kristallografiya, 25, 1980
|
|
2LH6
 
 | | X-RAY STRUCTURAL INVESTIGATION OF LEGHEMOGLOBIN. VI. STRUCTURE OF ACETATE-FERRILEGHEMOGLOBIN AT A RESOLUTION OF 2.0 ANGSTROMS (RUSSIAN) | | Descriptor: | LEGHEMOGLOBIN A (NICOTINATE MET), NICOTINIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Harutyunyan, E.H, Kuranova, I.P, Borisov, V.V, Sosfenov, N.I, Pavlovsky, A.G, Grebenko, A.I, Konareva, N.V. | | Deposit date: | 1982-04-23 | | Release date: | 1983-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structural Investigation of Leghemoglobin. Vi. Structure of Acetate-Ferrileghemoglobin at a Resolution of 2.0 Angstroms (Russian)
Kristallografiya, 25, 1980
|
|
2LH1
 
 | | X-RAY STRUCTURAL INVESTIGATION OF LEGHEMOGLOBIN. VI. STRUCTURE OF ACETATE-FERRILEGHEMOGLOBIN AT A RESOLUTION OF 2.0 ANGSTROMS (RUSSIAN) | | Descriptor: | ACETATE ION, LEGHEMOGLOBIN (ACETO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Harutyunyan, E.H, Kuranova, I.P, Borisov, V.V, Sosfenov, N.I, Pavlovsky, A.G, Grebenko, A.I, Konareva, N.V. | | Deposit date: | 1982-04-23 | | Release date: | 1983-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structural Investigation of Leghemoglobin. Vi. Structure of Acetate-Ferrileghemoglobin at a Resolution of 2.0 Angstroms (Russian)
Kristallografiya, 25, 1980
|
|
2LH5
 
 | | X-RAY STRUCTURAL INVESTIGATION OF LEGHEMOGLOBIN. VI. STRUCTURE OF ACETATE-FERRILEGHEMOGLOBIN AT A RESOLUTION OF 2.0 ANGSTROMS (RUSSIAN) | | Descriptor: | FLUORIDE ION, LEGHEMOGLOBIN (FLUORO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Harutyunyan, E.H, Kuranova, I.P, Borisov, V.V, Sosfenov, N.I, Pavlovsky, A.G, Grebenko, A.I, Konareva, N.V. | | Deposit date: | 1982-04-23 | | Release date: | 1983-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structural Investigation of Leghemoglobin. Vi. Structure of Acetate-Ferrileghemoglobin at a Resolution of 2.0 Angstroms (Russian)
Kristallografiya, 25, 1980
|
|
2N4J
 
 | |
2K3R
 
 | | Pfu Rpp21 structure and assignments | | Descriptor: | Ribonuclease P protein component 4, ZINC ION | | Authors: | Amero, C.D, Foster, M.P, Boomershine, W.P, Xu, Y. | | Deposit date: | 2008-05-15 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner.
Biochemistry, 47, 2008
|
|
2LH3
 
 | | X-RAY STRUCTURAL INVESTIGATION OF LEGHEMOGLOBIN. VI. STRUCTURE OF ACETATE-FERRILEGHEMOGLOBIN AT A RESOLUTION OF 2.0 ANGSTROMS (RUSSIAN) | | Descriptor: | CYANIDE ION, LEGHEMOGLOBIN (CYANO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Harutyunyan, E.H, Kuranova, I.P, Borisov, V.V, Sosfenov, N.I, Pavlovsky, A.G, Grebenko, A.I, Konareva, N.V. | | Deposit date: | 1982-04-23 | | Release date: | 1983-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structural Investigation of Leghemoglobin. Vi. Structure of Acetate-Ferrileghemoglobin at a Resolution of 2.0 Angstroms (Russian)
Kristallografiya, 25, 1980
|
|
7MJG
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
1VYW
 
 | | Structure of CDK2/Cyclin A with PNU-292137 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, N-(3-CYCLOPROPYL-1H-PYRAZOL-5-YL)-2-(2-NAPHTHYL)ACETAMIDE, ... | | Authors: | Pevarello, P, Brasca, M.G, Amici, R, Orsini, P, Traquandi, G, Corti, L, Piutti, C, Sansonna, P, Villa, M, Pierce, B.S, Pulici, M, Giordano, P, Martina, K, Fritzen, E.L, Nugent, R.A, Casale, E, Cameron, A, Ciomei, M, Roletto, F, Isacchi, A, Fogliatto, G, Pesenti, E, Pastori, W, Marsiglio, A, Leach, K.L, Clare, P.M, Fiorentini, F, Varasi, M, Vulpetti, A, Warpehoski, M.A. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2/Cyclin a as Antitumor Agents. Part 1. Lead Finding
J.Med.Chem., 47, 2004
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
1SRH
 
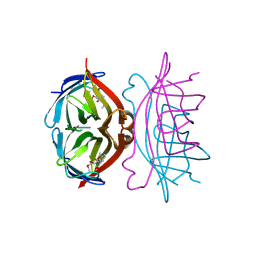 | |
6DV2
 
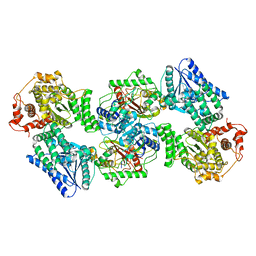 | | Crystal Structure of Human Mitochondrial Trifunctional Protein | | Descriptor: | Trifunctional enzyme subunit alpha, mitochondrial, Trifunctional enzyme subunit beta | | Authors: | Fu, Z, Xia, C, Battaile, K.P, Kim, J.P. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of human mitochondrial trifunctional protein, a fatty acid beta-oxidation metabolon.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1SPB
 
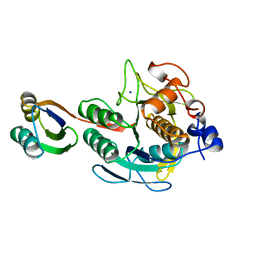 | | SUBTILISIN BPN' PROSEGMENT (77 RESIDUES) COMPLEXED WITH A MUTANT SUBTILISIN BPN' (266 RESIDUES). CRYSTAL PH 4.6. CRYSTALLIZATION TEMPERATURE 20 C DIFFRACTION TEMPERATURE-160 C | | Descriptor: | SODIUM ION, SUBTILISIN BPN', SUBTILISIN BPN' PROSEGMENT | | Authors: | Gallagher, D.T, Gilliland, G.L, Wang, L, Bryan, P.N. | | Deposit date: | 1995-06-21 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The prosegment-subtilisin BPN' complex: crystal structure of a specific 'foldase'.
Structure, 3, 1995
|
|
7U0H
 
 | | State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall model | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2022-02-18 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6L6F
 
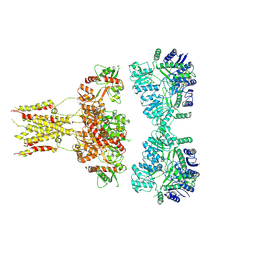 | | GluK3 receptor complex with UBP301 | | Descriptor: | Glutamate receptor ionotropic, kainate 3 | | Authors: | Kumar, J, Kumari, J, Burada, A.P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structural dynamics of the GluK3-kainate receptor neurotransmitter binding domains revealed by cryo-EM.
Int.J.Biol.Macromol., 149, 2020
|
|
7BL7
 
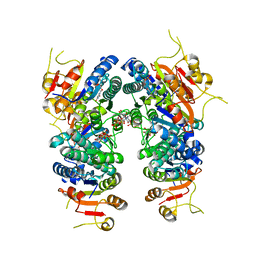 | | Crystal structure of UMPK from M. tuberculosis in complex with UDP and UTP (P21212 form) | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE, Uridylate kinase | | Authors: | Walter, P, Labesse, G, Haouz, A, Mechaly, A.E, Munier-Lehmann, H. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structural basis for the allosteric inhibition of UMP kinase from Gram-positive bacteria, a promising antibacterial target.
Febs J., 289, 2022
|
|
6HJP
 
 | |
