3G5Y
 
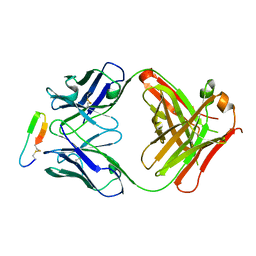 | |
1ZED
 
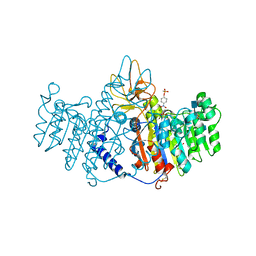 | | Alkaline phosphatase from human placenta in complex with p-nitrophenyl-phosphonate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, CALCIUM ION, ... | | Authors: | Llinas, P, Stura, E.A, Menez, A, Kiss, Z, Stigbrand, T, Millan, J.L, Le Du, M.H. | | Deposit date: | 2005-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Studies of Human Placental Alkaline Phosphatase in Complex with Functional Ligands.
J.Mol.Biol., 350, 2005
|
|
1EQW
 
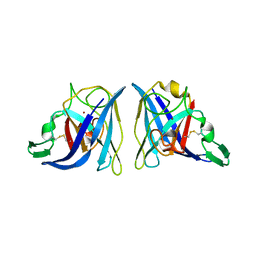 | | CRYSTAL STRUCTURE OF SALMONELLA TYPHIMURIUM CU,ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Pesce, A, Battistoni, A, Stroppolo, M.E, Polizio, F, Nardini, M, Kroll, J.S, Langford, P.R, O'Neill, P, Sette, M, Desideri, A, Bolognesi, M. | | Deposit date: | 2000-04-06 | | Release date: | 2000-09-08 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and crystallographic characterization of Salmonella typhimurium Cu,Zn superoxide dismutase coded by the sodCI virulence gene.
J.Mol.Biol., 302, 2000
|
|
5U4K
 
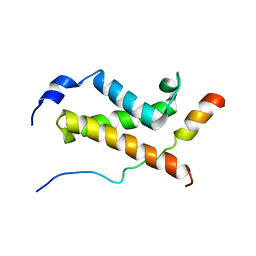 | | NMR structure of the complex between the KIX domain of CBP and the transactivation domain 1 of p65 | | Descriptor: | CREB-binding protein, Transcription factor p65 | | Authors: | Lecoq, L, Raiola, L, Chabot, P.R, Cyr, N, Arseneault, G, Omichinski, J.G. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
4BW2
 
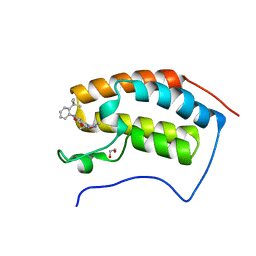 | | The first bromodomain of human BRD4 in complex with 3,5 dimethylisoxaxole ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-((2-(TERT-BUTYL)PHENYL)AMINO)-7-(3,5-dimethylisoxazol-4-yl)-1,8-naphthyridine-3-carboxylic acid, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Mirguet, O, Lamotte, Y, Bamborough, P, Delannee, D, Bouillot, A, Gellibert, F, Krysa, G, Lewis, A, Witherington, J, Huet, P, Dudit, Y, Trottet, L, Nicodeme, E. | | Deposit date: | 2013-06-29 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Naphthyridines as Novel Bet Family Bromodomain Inhibitors.
Chemmedchem, 9, 2014
|
|
4BRI
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with MG UMPPNP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]uridine, CHLORIDE ION, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
1QBT
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4R-(4ALPHA,5ALPHA,6ALPHA,7ALPHA)]-3,3'-{{TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-DIYL]BIS(METHYLENE)]BIS[N-1H-BENZIMIDAZOL-2-YLBENZAMIDE] | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cyclic urea amides: HIV-1 protease inhibitors with low nanomolar potency against both wild type and protease inhibitor resistant mutants of HIV.
J.Med.Chem., 40, 1997
|
|
5UIF
 
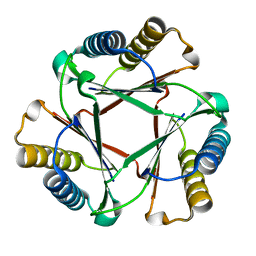 | | Crystal Structure of Native Ps01740 | | Descriptor: | Ps01740 | | Authors: | LeVieux, J, Baas, B.J, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2017-01-13 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Kinetic and structural characterization of a cis-3-Chloroacrylic acid dehalogenase homologue in Pseudomonas sp. UW4: A potential step between subgroups in the tautomerase superfamily.
Arch. Biochem. Biophys., 636, 2017
|
|
4CBU
 
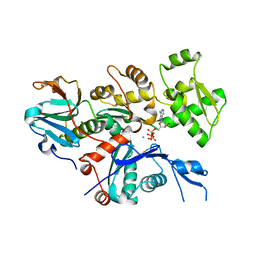 | | Crystal structure of Plasmodium falciparum actin I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-1, CALCIUM ION, ... | | Authors: | Vahokoski, J, Bhargav, S.P, Desfosses, A, Andreadaki, M, Kumpula, E.P, Ignatev, A, Munico Martinez, S, Lepper, S, Frischknecht, F, Siden-Kiamos, I, Sachse, C, Kursula, I. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Differences Explain Diverse Functions of Plasmodium Actins.
Plos Pathog., 10, 2014
|
|
5CP0
 
 | |
1QD7
 
 | | PARTIAL MODEL FOR 30S RIBOSOMAL SUBUNIT | | Descriptor: | CENTRAL FRAGMENT OF 16 S RNA, END FRAGMENT OF 16 S RNA, S15 RIBOSOMAL PROTEIN, ... | | Authors: | Clemons Jr, W.M, May, J.L.C, Wimberly, B.T, McCutcheon, J.P, Capel, M.S, Ramakrishnan, V. | | Deposit date: | 1999-07-09 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution.
Nature, 400, 1999
|
|
1ZNM
 
 | | A zinc finger with an artificial beta-turn, original sequence taken from the third zinc finger domain of the human transcriptional repressor protein YY1 (YING and YANG 1, a delta transcription factor), nmr, 34 structures | | Descriptor: | YY1, ZINC ION | | Authors: | Viles, J.H, Patel, S.U, Mitchell, J.B.O, Moody, C.M, Justice, D.E, Uppenbrink, J, Doyle, P.M, Harris, C.J, Sadler, P.J, Thornton, J.M. | | Deposit date: | 1997-11-20 | | Release date: | 1998-04-01 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Design, synthesis and structure of a zinc finger with an artificial beta-turn.
J.Mol.Biol., 279, 1998
|
|
4BRA
 
 | | Legionella pneumophila NTPDase1 crystal form II, closed, Mg AMPPNP complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, GLYCEROL, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
2H3M
 
 | | Crystal Structure of ZO-1 PDZ1 | | Descriptor: | SULFATE ION, Tight junction protein ZO-1 | | Authors: | Appleton, B.A, Zhang, Y, Wu, P, Yin, J.P, Hunziker, W, Skelton, N.J, Sidhu, S.S, Wiesmann, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparative structural analysis of the Erbin PDZ domain and the first PDZ domain of ZO-1. Insights into determinants of PDZ domain specificity.
J.Biol.Chem., 281, 2006
|
|
1UPX
 
 | | The crystal structure of the Hybrid Cluster Protein from Desulfovibrio desulfuricans containing molecules in the oxidized and reduced states. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FE4-S3 CLUSTER, HYDROXYLAMINE REDUCTASE, ... | | Authors: | Aragao, D, Macedo, S, Mitchell, E.P, Lindley, P.F. | | Deposit date: | 2003-10-14 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of the Hybrid Cluster Protein (Hcp) from Desulfovibrio Desulfuricans Atcc 27774 Containing Molecules in the Oxidized and Reduced States
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3BBH
 
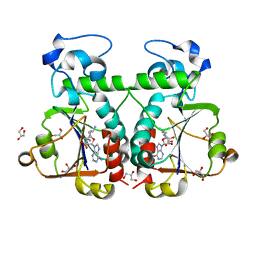 | | M. jannaschii Nep1 complexed with Sinefungin | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, SINEFUNGIN | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
7DFV
 
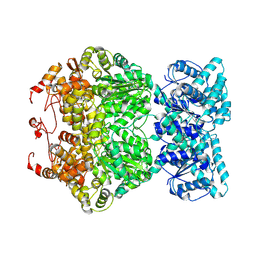 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | Descriptor: | NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-11-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
3P37
 
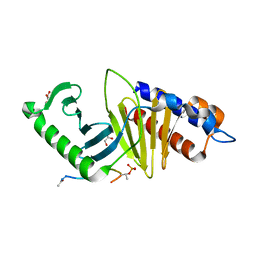 | |
1ZF0
 
 | | B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*AP*AP*CP*GP*G)-3', CALCIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5W3K
 
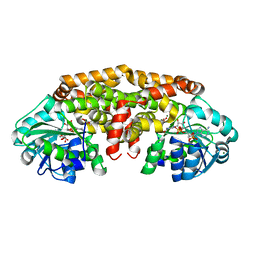 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex NADPH, Mg2+ and CPD | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Patel, K.M, Teran, D, Zheng, S, Kandale, A, Schembri, M, McGeary, R.P, Schenk, G, Guddat, L.W. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Crystal Structures of Staphylococcus aureus Ketol-Acid Reductoisomerase in Complex with Two Transition State Analogues that Have Biocidal Activity.
Chemistry, 23, 2017
|
|
3OQ2
 
 | | Structure of a CRISPR associated protein Cas2 from Desulfovibrio vulgaris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Samai, P, Smith, P, Shuman, S. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1YYK
 
 | | Crystal structure of RNase III from Aquifex Aeolicus complexed with double-stranded RNA at 2.5-angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1P5M
 
 | |
4BNP
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with isocitrate and magnesium(II) | | Descriptor: | ISOCITRATE DEHYDROGENASE [NADP], ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Miller, S.P, Goncalves, S, Matias, P.M, Dean, A.M. | | Deposit date: | 2013-05-16 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of a Transition State: Role of Lys100 in the Active Site of Isocitrate Dehydrogenase.
Chembiochem, 15, 2014
|
|
4BR2
 
 | | rat NTPDase2 in complex with Ca UMPPNP | | Descriptor: | 5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]uridine, CALCIUM ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-03 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
