5C7D
 
 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 17 | | Descriptor: | (2R)-4-[2-(6-chloro-2,3-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)-2-oxoethyl]-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
6JT8
 
 | | Crystal structure of 450-451_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
1AGB
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGRKKYKL-3R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGRKKYKL - 3R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
2OQ4
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli (E2Q) in complex with AP-site containing DNA substrate | | Descriptor: | 5'-D(*C*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*G*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Grollman, A.P, Shoahm, G. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site plasticity of endonuclease VIII: Conformational changes compensating for different substrates and mutations of critical residues
To be Published
|
|
5CCR
 
 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, POTASSIUM ION, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
6JT7
 
 | | Crystal structure of 452-453_deletion mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
3Q0V
 
 | | ETHR From mycobacterium tuberculosis in complex with compound bdm31369 | | Descriptor: | 4-methyl-1-{4-[3-(thiophen-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}pentan-1-one, HTH-type transcriptional regulator EthR | | Authors: | Flipo, M, Desroses, M, Dirie, B, Carette, X, Leroux, F, Lens, Z, Rucktooa, P, Leroux, F, Piveteau, C, Demirkaya, F, Locht, C, Villeret, V, Christophe, T, Jeon, H.K, Brodin, P, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis and optimization of new EthR inhibitors. A new alternative approach to fight tuberculosis by boosting ethionamide
To be Published
|
|
4R02
 
 | | yCP in complex with BSc4999 (alpha-Keto Phenylamide) | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-{(2S,3S)-1-[(2,4-dimethylphenyl)amino]-2-hydroxy-5-methyl-1-oxohexan-3-yl}-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Voss, C, Scholz, C, Knorr, S, Beck, P, Stein, M, Zall, A, Kuckelkorn, U, Kloetzel, P.-M, Groll, M, Hamacher, K, Schmidt, B. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | alpha-Keto Phenylamides as P1'-Extended Proteasome Inhibitors.
Chemmedchem, 9, 2014
|
|
5HU0
 
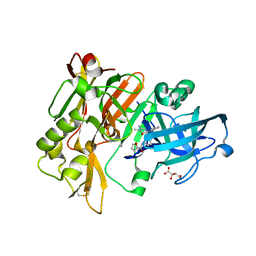 | | BACE1 in complex with 4-(3-(furan-2-carboxamido)phenyl)-1-methyl-5-oxo-4-phenylimidazolidin-2-iminium | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-{3-[(2E,4R)-2-imino-1-methyl-5-oxo-4-phenylimidazolidin-4-yl]phenyl}furan-2-carboxamide | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
5BXW
 
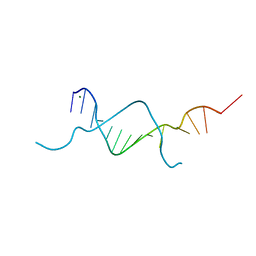 | |
5E3N
 
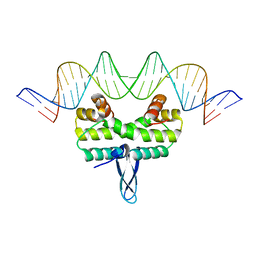 | |
6NW2
 
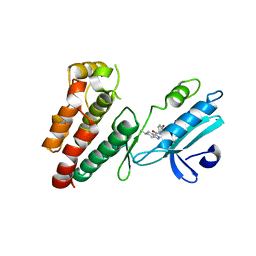 | | Structure of human RIPK1 kinase domain in complex with compound 11 | | Descriptor: | (5R)-5-methyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-4,5,6,7-tetrahydro-2H-indazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and selective inhibitors of receptor-interacting protein kinase 1 that lack an aromatic back pocket group.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
2Y37
 
 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 2-[(1R)-3-amino-1-phenyl-propoxy]-4-chloro-benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T.N, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2010-12-19 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1A7T
 
 | | METALLO-BETA-LACTAMASE WITH MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, METALLO-BETA-LACTAMASE, SODIUM ION, ... | | Authors: | Fitzgerald, P.M.D, Wu, J.K, Toney, J.H. | | Deposit date: | 1998-03-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unanticipated inhibition of the metallo-beta-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-A resolution.
Biochemistry, 37, 1998
|
|
7GZZ
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011184-001 | | Descriptor: | 2-[(4M)-4-(4-{[(1S)-1-(1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~-benzothiopyran-7-yl)-2-methylpropyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-1H-pyrazol-1-yl]-N-methylacetamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.166 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H12
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013388-001 | | Descriptor: | 7-[(1R)-2-methyl-1-{[(6M)-6-(1H-pyrazol-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}propyl]-3,4-dihydro-1lambda~6~-benzothiopyran-1,1(2H)-dione, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZ3
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000090-001 | | Descriptor: | (2S,3S)-N-(5,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-2-methyloxolane-3-carboxamide, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZG
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000620-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-[(1H-pyrrolo[3,2-b]pyridine-5-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZS
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0008340-001 | | Descriptor: | N-[(1R)-1-(4-methoxyphenyl)-2-(1H-tetrazol-5-yl)ethyl]-7H-pyrrolo[2,3-d]pyrimidine-4-carboxamide, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZM
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0008324-001 | | Descriptor: | N-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-D-valine, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZE
 
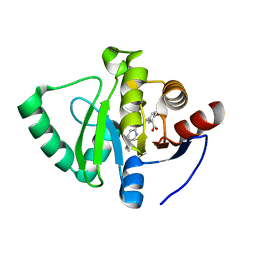 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000601-001 | | Descriptor: | (3R)-3-(4-bromophenyl)-3-{[5-(dimethylamino)pyridine-2-carbonyl]amino}propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZ1
 
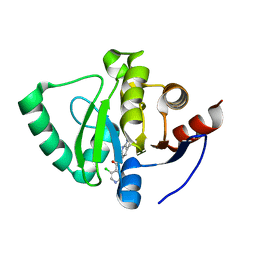 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000061-001 | | Descriptor: | 3-chloro-N-(1H-pyrazolo[3,4-b]pyridin-5-yl)pyridine-4-carboxamide, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZ2
 
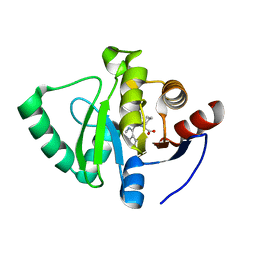 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000072-001 | | Descriptor: | (2R)-1-(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)azepane-2-carboxylic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.137 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZN
 
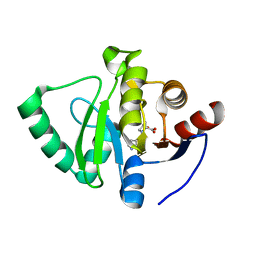 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0008351-001 | | Descriptor: | (4S)-4-hydroxy-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-D-proline, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZR
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0008273-001 | | Descriptor: | (3S)-3-[3-(methanesulfonamido)phenyl]-3-[(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)amino]propanoic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.455 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
