4Q3K
 
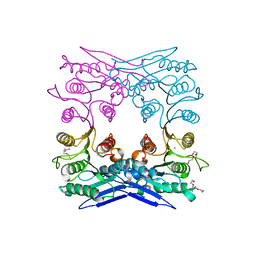 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
5U0G
 
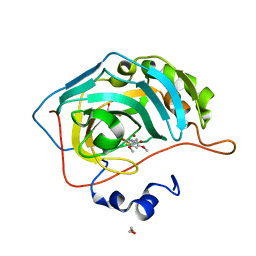 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-[(2,4-dimethoxyphenyl)methyl]-1,3-thiazolidine-2,4-dione, 1,2-ETHANEDIOL, Carbonic anhydrase 2, ... | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
6CME
 
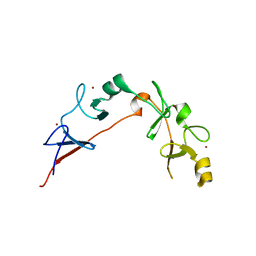 | | Structure of wild-type ISL2-LID in complex with LHX4-LIM1+2 | | Descriptor: | LIM/homeobox protein Lhx4,Insulin gene enhancer protein ISL-2, ZINC ION | | Authors: | Stokes, P.H, Silva, A, Guss, J.M, Matthews, J.M. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins, 87, 2019
|
|
5TYA
 
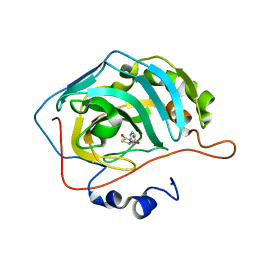 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-phenyl-1,3-thiazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-18 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
7Z3C
 
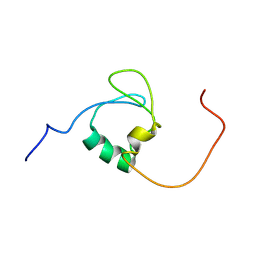 | | Solution structure of GltJ GYF domain from Myxococcus xanthus | | Descriptor: | Adventurous gliding motility protein X | | Authors: | Attia, B, My, L, Castaing, J.P, Le Guenno, H, Espinosa, L, Schmidt, V, Nouailler, M, Bornet, O, Elantak, L, Mignot, T. | | Deposit date: | 2022-03-02 | | Release date: | 2023-02-22 | | Last modified: | 2024-09-04 | | Method: | SOLUTION NMR | | Cite: | A molecular switch controls assembly of bacterial focal adhesions.
Sci Adv, 10, 2024
|
|
1MUU
 
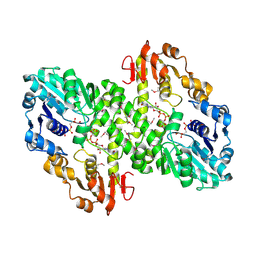 | | 2.0 A crystal structure of GDP-mannose dehydrogenase | | Descriptor: | GDP-mannose 6-dehydrogenase, GUANOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), P'-D-MANNOPYRANOSYL ESTER, ... | | Authors: | Snook, C.F, Tipton, P.A, Beamer, L.J. | | Deposit date: | 2002-09-24 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of GDP-mannose dehydrogenase: A key enzyme of alginate biosynthesis in P. aeruginosa
Biochemistry, 42, 2003
|
|
5U2O
 
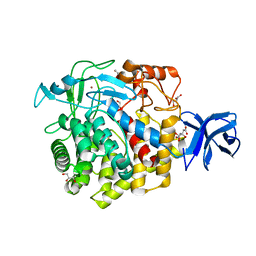 | | Crystal structure of Zn-binding triple mutant of GH family 9 endoglucanase J30 | | Descriptor: | CITRATE ANION, GLYCEROL, J30 CCH, ... | | Authors: | Ellinghaus, T.L, Pereira, J.H, McAndrew, R.P, Welner, D.H, Adams, P.D. | | Deposit date: | 2016-11-30 | | Release date: | 2018-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Engineering glycoside hydrolase stability by the introduction of zinc binding.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7BTQ
 
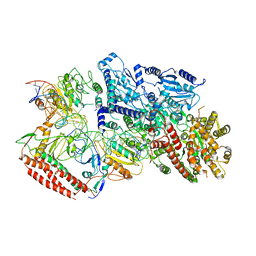 | | EcoR124I-DNA in the Restriction-Alleviation State | | Descriptor: | DNA (64-MER), Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
2YJG
 
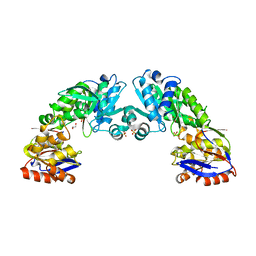 | | Structure of the lactate racemase apoprotein from Thermoanaerobacterium thermosaccharolyticum | | Descriptor: | 1,2-ETHANEDIOL, LACTATE RACEMASE APOPROTEIN, MAGNESIUM ION, ... | | Authors: | Declercq, J.P, Desguin, B, Soumillion, P, Hols, P. | | Deposit date: | 2011-05-19 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactate Racemase is a Nickel-Dependent Enzyme Activated by a Widespread Maturation System.
Nat.Commun., 5, 2014
|
|
7RU7
 
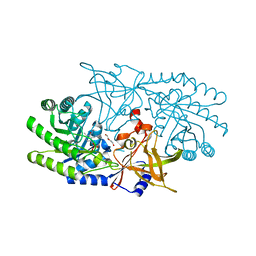 | | Crystal structure of BtrK, a decarboxylase involved in butirosin biosynthesis | | Descriptor: | DI(HYDROXYETHYL)ETHER, L-glutamyl-[BtrI acyl-carrier protein] decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Arenas, L.A.R, Paiva, F.C.R, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of BtrK, a decarboxylase involved in the (S)-4-amino-2-hydroxybutyrate (AHBA) formation during butirosin biosynthesis
J.Mol.Struct., 1267, 2022
|
|
4Q90
 
 | |
4Q9Q
 
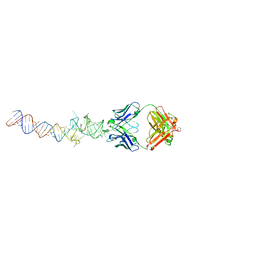 | | Crystal structure of an RNA aptamer bound to bromo-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3-bromobenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
7YZW
 
 | |
5XVX
 
 | | Crystal Structure of Aspergillus niger Glutamate Dehydrogenase Complexed With Alpha-ketoglutarate and NADPH | | Descriptor: | 2-OXOGLUTARIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Prakash, P, Punekar, N.S, Bhaumik, P. | | Deposit date: | 2017-06-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the catalytic mechanism and alpha-ketoglutarate cooperativity of glutamate dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
4QBE
 
 | | The second sphere residue T263 is important for function and activity of PTP1B through modulating WPD loop | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xiao, P, Wang, X, Wang, H.M, Fu, X.L, Cui, F.A, Yu, X, Bi, W.X. | | Deposit date: | 2014-05-08 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | The second-sphere residue T263 is important for the function and catalytic activity of PTP1B via interaction with the WPD-loop
Int.J.Biochem.Cell Biol., 57C, 2014
|
|
6SNI
 
 | | Cryo-EM structure of nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab | | Descriptor: | 6AG9-Fab heavy chain, 6AG9-Fab light chain, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
4QBY
 
 | | yCP in complex with BOC-ALA-ALA-ALA-CHO | | Descriptor: | BOC-ALA-ALA-ALA-CHO, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Arciniega, M, Beck, P, Lange, O, Groll, M, Huber, R. | | Deposit date: | 2014-05-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential global structural changes in the core particle of yeast and mouse proteasome induced by ligand binding.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1A2U
 
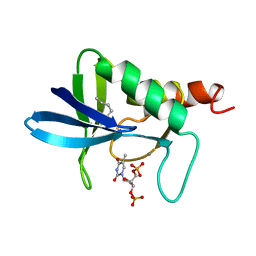 | | STAPHYLOCOCCAL NUCLEASE, V23C VARIANT, COMPLEX WITH 1-N-BUTANE THIOL AND 3',5'-THYMIDINE DIPHOSPHATE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1998-01-11 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mobile unnatural amino acid side chains in the core of staphylococcal nuclease.
Protein Sci., 5, 1996
|
|
7Z1I
 
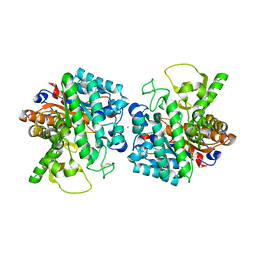 | | Plant myrosinase TGG1 from Arabidopsis thaliana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gao, Y, Farmer, E, Jimenez-Sandoval, P, Santiago, J. | | Deposit date: | 2022-02-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Plant myrosinase TGG1 from Arabidopsis thaliana
To Be Published
|
|
7RUJ
 
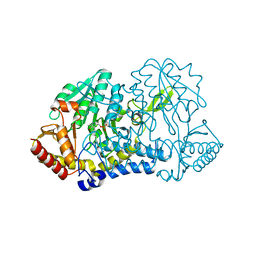 | | E. coli cysteine desulfurase SufS N99A | | Descriptor: | CHLORIDE ION, Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Gogar, R, Frantom, P.A. | | Deposit date: | 2021-08-17 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The beta-latch structural element of the SufS cysteine desulfurase mediates active site accessibility and SufE transpersulfurase positioning.
J.Biol.Chem., 299, 2023
|
|
1WV5
 
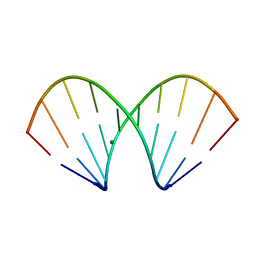 | | X-ray structure of the A-decamer GCGTATACGC with a single 2'-o-butyl thymine in place of T6, Mg-form | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(2BT)P*AP*CP*GP*C)-3', MAGNESIUM ION | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharam, M. | | Deposit date: | 2004-12-11 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the influence of stereoelectronic effects on the biophysical properties of oligonucleotides: comprehensive analysis of the RNA affinity, nuclease resistance, and crystal structure of ten 2'-O-ribonucleic acid modifications.
Biochemistry, 44, 2005
|
|
7RW3
 
 | | E. coli cysteine desulfurase SufS N99D | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Gogar, R, Frantom, P.A. | | Deposit date: | 2021-08-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The beta-latch structural element of the SufS cysteine desulfurase mediates active site accessibility and SufE transpersulfurase positioning.
J.Biol.Chem., 299, 2023
|
|
5C0R
 
 | | Crystal Structure of a Generation 3 Influenza Hemagglutinin Stabilized Stem Complexed with the Broadly Neutralizing Antibody C179 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C179 Fab heavy chain, ... | | Authors: | Boyington, J.C, kwong, P.D, Nabel, G.J, Mascola, J.R. | | Deposit date: | 2015-06-12 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection.
Nat. Med., 21, 2015
|
|
7Z6T
 
 | | Aspergillus clavatus M36 protease without the propeptide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Extracellular metalloproteinase mep, ... | | Authors: | Wilkens, C, Qiu, J, Meyer, A.S, Morth, J.P. | | Deposit date: | 2022-03-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Aspergillus clavatus M36 protease without the propeptide
To Be Published
|
|
3GZC
 
 | | Structure of human selenocysteine lyase | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Selenocysteine lyase | | Authors: | Collins, R, Hogbom, M, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Karlberg, T, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
