6IDS
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN D35N mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
2FP7
 
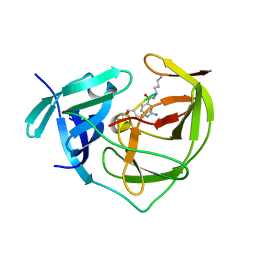 | |
1R81
 
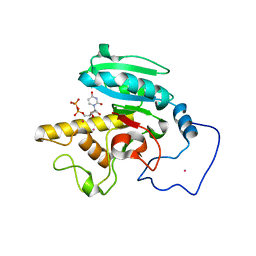 | | Glycosyltransferase A in complex with 3-amino-acceptor analog inhibitor and uridine diphosphate-N-acetyl-galactose | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MERCURY (II) ION, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE, ... | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
3RYG
 
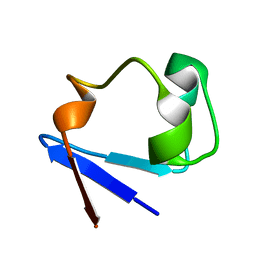 | | 128 hours neutron structure of perdeuterated rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1R8O
 
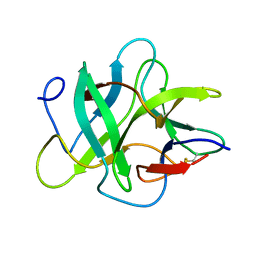 | | Crystal structure of an unusual Kunitz-type trypsin inhibitor from Copaifera langsdorffii seeds | | Descriptor: | Kunitz trypsin inhibitor | | Authors: | Krauchenco, S, Nagem, R.A.P, da Silva, J.A, Marangoni, S, Polikarpov, I. | | Deposit date: | 2003-10-27 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Three-dimensional structure of an unusual Kunitz (STI) type trypsin inhibitor from Copaifera langsdorffii.
Biochimie, 86, 2004
|
|
6E18
 
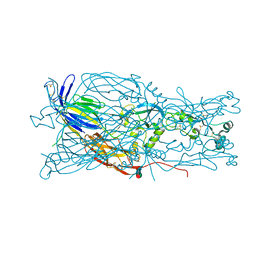 | | Crystal structure of Chlamydomonas reinhardtii HAP2 ectodomain provides structural insights of functional loops in green algae. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Baquero, E, Legrand, P, Rey, F.A. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Species-Specific Functional Regions of the Green Alga Gamete Fusion Protein HAP2 Revealed by Structural Studies.
Structure, 27, 2019
|
|
1RAV
 
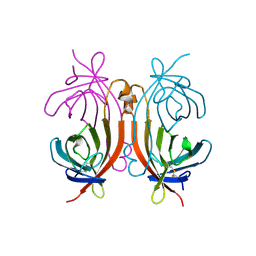 | | RECOMBINANT AVIDIN | | Descriptor: | AVIDIN | | Authors: | Rosano, C, Arosio, P, Bolognesi, M. | | Deposit date: | 1998-03-27 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical characterization and crystal structure of a recombinant hen avidin and its acidic mutant expressed in Escherichia coli.
Eur.J.Biochem., 256, 1998
|
|
5XTO
 
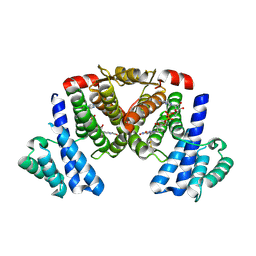 | |
5E7B
 
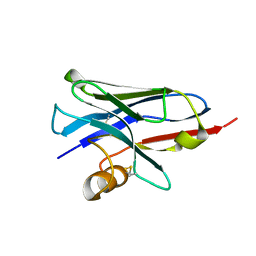 | | Structure of a nanobody (vHH) from camel against phage Tuc2009 RBP (BppL, ORF53) | | Descriptor: | nanobody nano-L06 | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-12 | | Release date: | 2015-12-30 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
1IHU
 
 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE IN COMPLEX WITH MG-ADP-ALF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ARSENICAL PUMP-DRIVING ATPASE, ... | | Authors: | Zhou, T, Radaev, S, Rosen, B.P, Gatti, D.L. | | Deposit date: | 2001-04-20 | | Release date: | 2001-09-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
3LVP
 
 | | Crystal structure of bisphosphorylated IGF1-R Kinase domain (2P) in complex with a bis-azaindole inhibitor | | Descriptor: | 3-(4-chloro-1H-pyrrolo[2,3-b]pyridin-2-yl)-5,6-dimethoxy-1-methyl-1H-pyrrolo[3,2-b]pyridine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Insulin-like growth factor 1 receptor, ... | | Authors: | Maignan, S, Marquette, J.P, Guilloteau, J.P. | | Deposit date: | 2010-02-22 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design of Potent IGF1-R Inhibitors Related to Bis-azaindoles
Chem.Biol.Drug Des., 76, 2010
|
|
5DW7
 
 | | Crystal structure of the unliganded geosmin synthase N-terminal domain from Streptomyces coelicolor | | Descriptor: | Germacradienol/geosmin synthase | | Authors: | Lombardi, P.M, Harris, G.G, Pemberton, T.A, Matsui, T, Weiss, T.M, Cole, K.E, Koksal, M, Murphy, F.V, Vedula, L.S, Chou, W.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2015-09-22 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural Studies of Geosmin Synthase, a Bifunctional Sesquiterpene Synthase with alpha alpha Domain Architecture That Catalyzes a Unique Cyclization-Fragmentation Reaction Sequence.
Biochemistry, 54, 2015
|
|
5XCK
 
 | | Crystal structure of Vps29 double mutant (D62A/H86A) from Entamoeba histolytica | | Descriptor: | Vacuolar protein sorting-associated protein 29 | | Authors: | Srivastava, V.K, Yadav, R, Tomar, P, Gourinath, S, Datta, S. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic characterization of metal binding in Vps29 from Entamoeba histolytica: implication in retromer function.
Mol. Microbiol., 106, 2017
|
|
1RLI
 
 | |
5XWA
 
 | | Crystal Structure of Porcine pancreatic trypsin with tripeptide inhibitor, PRY, at pH 10 | | Descriptor: | Acetylated-Pro-Arg-Tyr Inhibitor, CALCIUM ION, Trypsin | | Authors: | Saikhedkar, N.S, Bhoite, A.S, Giri, A.P, Kulkarni, K.A. | | Deposit date: | 2017-06-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tripeptides derived from reactive centre loop of potato type II protease inhibitors preferentially inhibit midgut proteases of Helicoverpa armigera.
Insect Biochem. Mol. Biol., 95, 2018
|
|
4KXX
 
 | | Human transketolase in covalent complex with donor ketose D-sedoheptulose-7-phosphate | | Descriptor: | (2R,3R,4S,5R,6S)-2,3,4,5,6,7-hexahydroxyheptyl dihydrogen phosphate, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
2FVL
 
 | | Crystal structure of human 3-alpha hydroxysteroid/dihydrodiol dehydrogenase (AKR1C4) complexed with NADP+ | | Descriptor: | Aldo-keto reductase family 1, member C4, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ugochukwu, E, Smee, C, Guo, K, Lukacik, P, Kavanagh, K, Debreczeni, J.E, von Delft, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human 3-alpha hydroxysteroid/dihydrodiol dehydrogenase (AKR1C4) complexed with NADP+
To be Published
|
|
3S8H
 
 | | Structure of dihydrodipicolinate synthase complexed with 3-Hydroxypropanoic acid(HPA)at 2.70 A resolution | | Descriptor: | 3-HYDROXY-PROPANOIC ACID, Dihydrodipicolinate synthase | | Authors: | Kumar, M, Kaur, N, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-05-28 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of dihydrodipicolinate synthase complexed with 3-Hydroxypropanoic acid(HPA)at 2.70 A resolution
TO BE PUBLISHED
|
|
6IGP
 
 | | Crystal structure of S9 peptidase (inactive state)from Deinococcus radiodurans R1 in P212121 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
3LTV
 
 | | Mouse-human sod1 chimera | | Descriptor: | Superoxide dismutase [Cu-Zn],Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Seetharaman, S.V, Taylor, A.B, Hart, P.J. | | Deposit date: | 2010-02-16 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Structures of mouse SOD1 and human/mouse SOD1 chimeras.
Arch.Biochem.Biophys., 503, 2010
|
|
6EGS
 
 | | Crystal structure of the GalNAc-T2 F104S mutant in complex with UDP-GalNAc | | Descriptor: | MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | de las Rivas, M, Coelho, H, Diniz, A, Lira-Navarrete, E, Jimenez-Barbero, J, Schjoldager, K.T, Bennett, E.P, Vakhrushev, S.Y, Clausen, H, Corzana, F, Marcelo, F, Hurtado-Guerrero, R. | | Deposit date: | 2017-09-12 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Analysis of a GalNAc-T2 Mutant Reveals an Induced-Fit Catalytic Mechanism for GalNAc-Ts.
Chemistry, 24, 2018
|
|
5E1W
 
 | |
1R1H
 
 | | STRUCTURAL ANALYSIS OF NEPRILYSIN WITH VARIOUS SPECIFIC AND POTENT INHIBITORS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[3-[(1-AMINOETHYL)(HYDROXY)PHOSPHORYL]-2-(1,1'-BIPHENYL-4-YLMETHYL)PROPANOYL]ALANINE, Neprilysin, ... | | Authors: | Oefner, C, Roques, B.P, Fournie-Zaluski, M.C, Dale, G.E. | | Deposit date: | 2003-09-24 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of neprilysin with various specific and potent inhibitors.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ESQ
 
 | | CRYSTAL STRUCTURE OF THIAZOLE KINASE MUTANT (C198S) WITH ATP AND THIAZOLE PHOSPHATE. | | Descriptor: | 4-METHYL-5-HYDROXYETHYLTHIAZOLE PHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, HYDROXYETHYLTHIAZOLE KINASE, ... | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-04-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
6AAA
 
 | |
