5ION
 
 | |
2MPI
 
 | | Solution structure of B24G insulin | | Descriptor: | insulin chain A, insulin chain B | | Authors: | Yang, Y, Wickramasinghe, N.P, Hua, Q, Weiss, M.A. | | Deposit date: | 2014-05-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protective hinge in insulin opens to enable its receptor engagement.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4R5G
 
 | | Crystal structure of the DnaK C-terminus with the inhibitor PET-16 | | Descriptor: | Chaperone protein DnaK, triphenyl(phenylethynyl)phosphonium | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4501 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
3I8C
 
 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDR21A | | Descriptor: | DNA damage-binding protein 1, WD repeat-containing protein 21A | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4LBS
 
 | | Crystal structure of human AR complexed with NADP+ and {2-[(4-bromo-2,6-difluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {2-[(4-bromo-2,6-difluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.76 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
2J1Q
 
 | | Crystal structure of Trypanosoma cruzi arginine kinase | | Descriptor: | ARGININE KINASE, GLYCEROL | | Authors: | Fernandez, P, Haouz, A, Pereira, C.A, Aguilar, C, Alzari, P.M. | | Deposit date: | 2006-08-15 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Arginine Kinase.
Proteins, 69, 2007
|
|
2N1O
 
 | |
1W9K
 
 | | Dictyostelium discoideum Myosin II motor domain S456E with bound MgADP-BeFx | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Morris, C.A, Coureux, P.-D, Wells, A.L, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2004-10-13 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Function Analysis of Myosin II Backdoor Mutants
To be Published
|
|
1R3S
 
 | | Uroporphyrinogen Decarboxylase single mutant D86G in complex with coproporphyrinogen-I | | Descriptor: | COPROPORPHYRINOGEN I, Uroporphyrinogen Decarboxylase | | Authors: | Phillips, J.D, Whitby, F.G, Kushner, J.P, Hill, C.P. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for tetrapyrrole coordination by uroporphyrinogen decarboxylase
Embo J., 22, 2003
|
|
4IIK
 
 | | Legionella pneumophila effector | | Descriptor: | Adenosine monophosphate-protein hydrolase SidD, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tascon, I, Chen, Y, Neunuebel, M.R, Rojas, A.L, Machner, M.P, Hierro, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Rab1 De-AMPylation by the Legionella pneumophila Effector SidD
Plos Pathog., 9, 2013
|
|
4IIP
 
 | | Legionella pneumophila effector | | Descriptor: | Adenosine monophosphate-protein hydrolase SidD, CHLORIDE ION, GLYCEROL | | Authors: | Tascon, I, Chen, Y, Neunuebel, M.R, Rojas, A.L, Machner, M.P, Hierro, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Rab1 De-AMPylation by the Legionella pneumophila Effector SidD
Plos Pathog., 9, 2013
|
|
1W0G
 
 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, METYRAPONE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
7W9H
 
 | | Crystal structure of the receiver domain of the transcription regulator FleR from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, CALCIUM ION, Response regulator protein FleR | | Authors: | Sahoo, P.K, Sheenu, n, Jain, D. | | Deposit date: | 2021-12-09 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | REC domain stabilizes the active heptamer of sigma 54 -dependent transcription factor, FleR from Pseudomonas aeruginosa.
Iscience, 26, 2023
|
|
4BDT
 
 | | Human acetylcholinesterase in complex with huprine W and fasciculin 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Nachon, F, Carletti, E, Ronco, C, Trovaslet, M, Nicolet, Y, Jean, L, Renard, P.-Y. | | Deposit date: | 2012-10-06 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Crystal structures of human cholinesterases in complex with huprine W and tacrine: elements of specificity for anti-Alzheimer's drugs targeting acetyl- and butyryl-cholinesterase.
Biochem. J., 453, 2013
|
|
1W04
 
 | | Isopenicillin N Synthase Aminoadipoyl-Cysteinyl-Glycine-Fe-NO Complex | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-GLYCINE, FE (II) ION, HYDROXYAMINE, ... | | Authors: | Long, A.J, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-06-01 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Truncated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-Glycine and Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D- Alanine
Biochemistry, 44, 2005
|
|
1W0F
 
 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, PROGESTERONE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
7K5H
 
 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-66 | | Descriptor: | 4-{[3-(3-chloro-5-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
2IUC
 
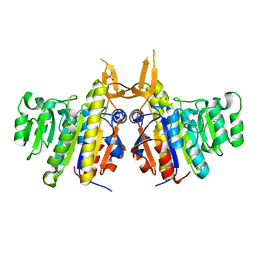 | | Structure of alkaline phosphatase from the Antarctic bacterium TAB5 | | Descriptor: | ALKALINE PHOSPHATASE, CACODYLATE ION, MAGNESIUM ION, ... | | Authors: | Wang, E, Koutsioulis, D, Leiros, H.K.S, Andersen, O.A, Bouriotis, V, Hough, E, Heikinheimo, P. | | Deposit date: | 2006-06-01 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Alkaline Phosphatase from the Antarctic Bacterium Tab5.
J.Mol.Biol., 366, 2007
|
|
1W7G
 
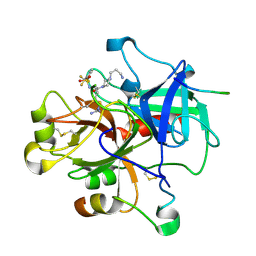 | | Alpha-thrombin complex with sulfated hirudin (residues 54-65) and L- Arginine template inhibitor CS107 | | Descriptor: | HIRUDIN I, N-{(1S)-1-{[4-(3-AMINOPROPYL)PIPERAZIN-1-YL]CARBONYL}-4-[(DIAMINOMETHYLENE)AMINO]BUTYL}-3-(TRIFLUOROMETHYL)BENZENESULFONAMIDE, THROMBIN | | Authors: | Remiche, J, Sauvage, E, Herman, R, Charlier, P. | | Deposit date: | 2004-09-02 | | Release date: | 2006-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, Synthesis and Evaluation of Graftable Thrombin Inhibitors for the Preparation of Blood-Compatible Polymer Materials.
Org.Biomol.Chem., 3, 2005
|
|
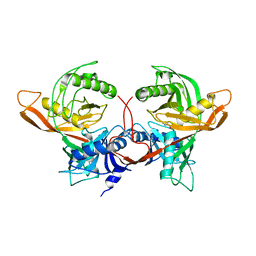
1W62
 
 | |
1U6C
 
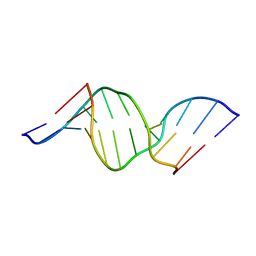 | | The NMR-derived solution structure of the (2S,3S)-N6-(2,3,4-trihydroxybutyl)-2'-deoxyadenosyl DNA adduct of butadiene diol epoxide | | Descriptor: | (2S,3S)-N6-(2,3,4-trihydroxybutyl)-2'-deoxyadenosyl DNA adduct of butadiene diol epoxide | | Authors: | Scholdberg, T.A, Nechev, L.N, Merritt, W.K, Harris, T.M, Harris, C.M, Lloyd, R.S, Stone, M.P. | | Deposit date: | 2004-07-29 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a site specific major groove (2S,3S)-N6-(2,3,4-trihydroxybutyl)-2'-deoxyadenosyl DNA adduct of butadiene diol epoxide.
Chem.Res.Toxicol., 17, 2004
|
|
1W06
 
 | | Isopenicillin N Synthase Aminoadipoyl-Cysteinyl-Alanine-Fe NO Complex | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-ALANINE, FE (II) ION, HYDROXYAMINE, ... | | Authors: | Long, A.J, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-06-01 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with the Truncated Substrate Analogues Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-Glycine and Delta-(L-Alpha-Aminoadipoyl)-L-Cysteinyl-D- Alanine
Biochemistry, 44, 2005
|
|
1FEC
 
 | |
7K4Z
 
 | | Crystal structure of Kemp Eliminase HG3.17 in complex with the transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, PENTAETHYLENE GLYCOL | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
1TS5
 
 | | I140T MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
