3FZF
 
 | | Crystal Structure of Hsc70/Bag1 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BAG family molecular chaperone regulator 1, Heat shock cognate 71 kDa protein | | Authors: | Dokurno, P, Williamson, D.S, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design
J.Med.Chem., 52, 2009
|
|
4NPO
 
 | | Crystal structure of protein with unknown function from Deinococcus radiodurans at P61 spacegroup | | Descriptor: | ACETATE ION, ACETYL GROUP, GLYCEROL, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korgenevsky, D.A, Shabalin, I.G, Shumilin, I.A, Dorovatovsky, P.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure of protein with unknown function at P61 spacegroup
To be Published
|
|
4NQM
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a triazolo-phthalazine ligand | | Descriptor: | 2-chloro-N-[5-(3-methyl[1,2,4]triazolo[3,4-a]phthalazin-6-yl)-2-(morpholin-4-yl)phenyl]benzenesulfonamide, Bromodomain-containing protein 4, SUCCINIC ACID | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-25 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with a triazolo-phthalazine ligand
To be Published
|
|
4NS4
 
 | | Crystal structure of cold-active estarase from Psychrobacter cryohalolentis K5T | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Boyko, K.M, Petrovskaya, L.E, Gorbacheva, M.A, Korgenevsky, D.A, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2013-11-28 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimentional structure of an esterse from Psychrobacter cryohalolentis K5T provides clues to unusual thermostability of a cold-active enzyme
To be Published
|
|
1AJ0
 
 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX OF E. COLI DIHYDROPTEROATE SYNTHASE | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, DIHYDROPTEROATE SYNTHASE, SULFANILAMIDE, ... | | Authors: | Achari, A, Somers, D.O, Champness, J.N, Bryant, P.K, Rosemond, J, Stammers, D.K. | | Deposit date: | 1997-05-14 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase.
Nat.Struct.Biol., 4, 1997
|
|
2EBA
 
 | |
2ES4
 
 | | Crystal structure of the Burkholderia glumae lipase-specific foldase in complex with its cognate lipase | | Descriptor: | CALCIUM ION, IODIDE ION, Lipase, ... | | Authors: | Pauwels, K, Wyns, L, Tommassen, J, Savvides, S.N, Van Gelder, P. | | Deposit date: | 2005-10-25 | | Release date: | 2006-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a membrane-based steric chaperone in complex with its lipase substrate.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1A2O
 
 | | STRUCTURAL BASIS FOR METHYLESTERASE CHEB REGULATION BY A PHOSPHORYLATION-ACTIVATED DOMAIN | | Descriptor: | CHEB METHYLESTERASE | | Authors: | Djordjevic, S, Goudreau, P.N, Xu, Q, Stock, A.M, West, A.H. | | Deposit date: | 1998-01-06 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4W2H
 
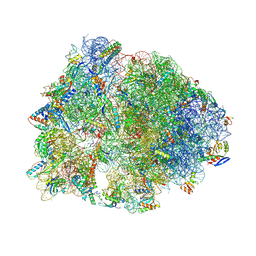 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pactamycin (co-crystallized), mRNA and deacylated tRNA in the P site | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
1B2V
 
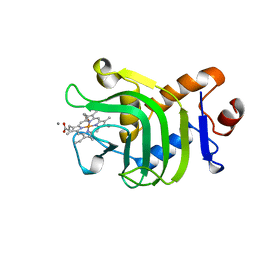 | | HEME-BINDING PROTEIN A | | Descriptor: | CALCIUM ION, PROTEIN (HEME-BINDING PROTEIN A), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnoux, P, Haser, R, Izadi, N, Lecroisey, A, Wandersma, N.C, Czjzek, M. | | Deposit date: | 1998-12-01 | | Release date: | 1999-06-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of HasA, a hemophore secreted by Serratia marcescens.
Nat.Struct.Biol., 6, 1999
|
|
8Y6O
 
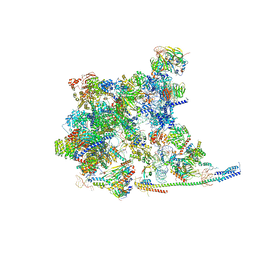 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U11 and tri-snRNP part | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Centrosomal AT-AC splicing factor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
6ZJ6
 
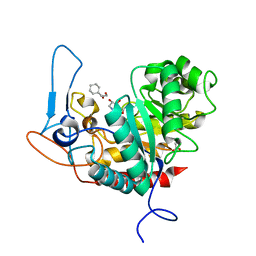 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with cyclohexylmethyl-Glc-1,3-isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ACETATE ION, ... | | Authors: | Thompson, A.J, Sobala, L.F, Fernandes, P.Z, Hakki, Z, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZHB
 
 | | 3D electron diffraction structure of bovine insulin | | Descriptor: | Insulin, ZINC ION | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-22 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.25 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5VIZ
 
 | |
3SQR
 
 | | Crystal structure of laccase from Botrytis aclada at 1.67 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, SULFATE ION, ... | | Authors: | Osipov, E.M, Polyakov, K.M, Tikhonova, T.V, Dorovatovsky, P.V, Ludwig, R, Kittl, R, Shleev, S.V, Popov, V.O. | | Deposit date: | 2011-07-06 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5VKJ
 
 | | Crystal structure of human CD22 Ig domains 1-3 | | Descriptor: | B-cell receptor CD22, GLYCEROL, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Julien, J.P, Ereno-Orbea, J, Sicard, T. | | Deposit date: | 2017-04-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun, 8, 2017
|
|
3STW
 
 | |
3T2N
 
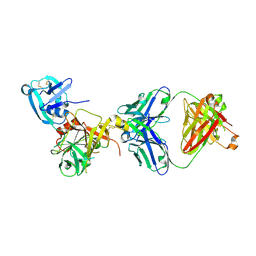 | | Human hepsin protease in complex with the Fab fragment of an inhibitory antibody | | Descriptor: | Antibody, Fab fragment, Heavy Chain, ... | | Authors: | Koschubs, T, Dengl, S, Duerr, H, Kaluza, K, Georges, G, Hartl, C, Jennewein, S, Lanzendoerfer, M, Auer, J, Stern, A, Huang, K.-S, Kostrewa, D, Ries, S, Hansen, S, Kohnert, U, Cramer, P, Mundigl, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-28 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Allosteric antibody inhibition of human hepsin protease.
Biochem.J., 442, 2012
|
|
4RX4
 
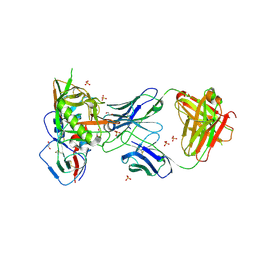 | | Crystal structure of VH1-46 germline-derived CD4-binding site-directed antibody 8ANC134 in complex with HIV-1 clade A Q842.d12 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC134 Heavy chain, ... | | Authors: | Zhou, T, Acharya, P, Moquin, S, Kwong, P.D. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-01 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell(Cambridge,Mass.), 161, 2015
|
|
4RWY
 
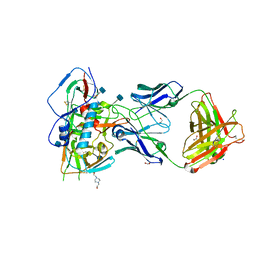 | | Crystal structure of VH1-46 germline-derived CD4-binding site-directed antibody 8ANC131 in complex with HIV-1 clade B YU2 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Acharya, P, Luongo, T.S, Kwong, P.D. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell(Cambridge,Mass.), 161, 2015
|
|
7RFA
 
 | |
3SU1
 
 | | Crystal structure of NS3/4A protease variant D168A in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, Genome polyprotein, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SW6
 
 | |
3TBA
 
 | | Structure of Yeast Ribonucleotide Reductase 1 Q288A with dGTP and ADP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C. | | Deposit date: | 2011-08-05 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Role of Arginine 293 and Glutamine 288 in Communication between Catalytic and Allosteric Sites in Yeast Ribonucleotide Reductase.
J.Mol.Biol., 419, 2012
|
|
3SU6
 
 | | Crystal structure of NS3/4A protease variant A156T in complex with vaniprevir | | Descriptor: | (5R,7S,10S)-10-tert-butyl-N-{(1R,2R)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethylcyclopropyl}-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19-dodecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosine-7(3H)-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
