3RQ4
 
 | | Crystal structure of suppressor of variegation 4-20 homolog 2 | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SUV420H2, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Zeng, H, Tempel, W, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-27 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2.
Febs Lett., 587, 2013
|
|
7F71
 
 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) with peptidoglycan sugar moiety and glutamate | | Descriptor: | GLUTAMIC ACID, GLYCEROL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2021-06-26 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Allosteric cooperation in beta-lactam binding to a non-classical transpeptidase.
Elife, 11, 2022
|
|
5EXO
 
 | | Crystal structure of Human galectin-3 CRD in complex with methyl 2-O-acetyl-3-O-(2H-chromene-3-yl-methyl)-a-D-galactopyranoside inhibitor | | Descriptor: | CHLORIDE ION, Galectin-3, [(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2-methoxy-5-oxidanyl-4-[(2-oxidanylidenechromen-3-yl)methoxy]oxan-3-yl] ethanoate | | Authors: | Collins, P.M, Blanchard, H. | | Deposit date: | 2015-11-23 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | A Selective Galactose-Coumarin-Derived Galectin-3 Inhibitor Demonstrates Involvement of Galectin-3-glycan Interactions in a Pulmonary Fibrosis Model.
J.Med.Chem., 59, 2016
|
|
2N2E
 
 | | NMR solution structure of the C-terminal domain of NisI, a lipoprotein from Lactococcus lactis which confers immunity against nisin | | Descriptor: | Nisin immunity protein | | Authors: | Hacker, C, Christ, N.A, Korn, S, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Entian, K, Woehnert, J. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Lantibiotic Immunity Protein NisI and Its Interactions with Nisin.
J.Biol.Chem., 290, 2015
|
|
1AKS
 
 | | CRYSTAL STRUCTURE OF THE FIRST ACTIVE AUTOLYSATE FORM OF THE PORCINE ALPHA TRYPSIN | | Descriptor: | ALPHA TRYPSIN, CALCIUM ION | | Authors: | Johnson, A, Krishnaswamy, S, Sundaram, P.V, Pattabhi, V. | | Deposit date: | 1996-07-24 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure at 1.8 A resolution of an active autolysate form of porcine alpha-trysoin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
7KXS
 
 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2020-12-04 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5J96
 
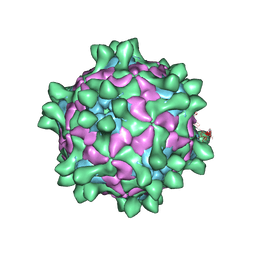 | | Crystal structure of Slow Bee Paralysis Virus at 3.4A resolution | | Descriptor: | Genome polyprotein, VP1, VP2 | | Authors: | Kalynych, S, Levdansky, Y, Palkova, L, Plevka, P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Virion Structure of Iflavirus Slow Bee Paralysis Virus at 2.6-Angstrom Resolution.
J.Virol., 90, 2016
|
|
1OCJ
 
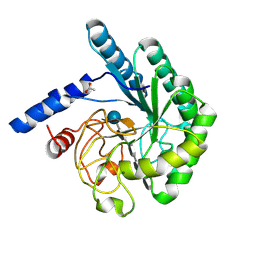 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a THIOPENTASACCHARIDE at 1.3 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1A1X
 
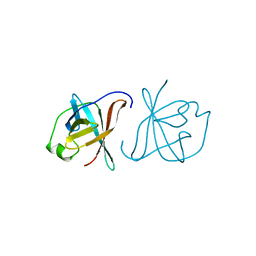 | | CRYSTAL STRUCTURE OF MTCP-1 INVOLVED IN T CELL MALIGNANCIES | | Descriptor: | HMTCP-1 | | Authors: | Fu, Z.Q, Dubois, G.C, Song, S.P, Kulikovskaya, I, Virgilio, L, Rothstein, J, Croce, C.M, Weber, I.T, Harrison, R.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MTCP-1: implications for role of TCL-1 and MTCP-1 in T cell malignancies.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6RNF
 
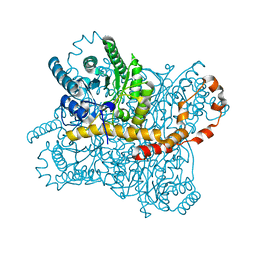 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 30 ms timepoint | | Descriptor: | MAGNESIUM ION, Xylose isomerase, alpha-D-glucopyranose | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-05-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
1O2K
 
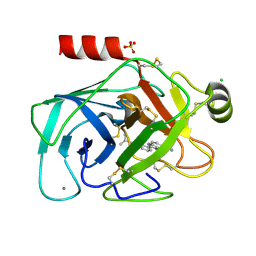 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-6-ISOBUTOXYBENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
6EEY
 
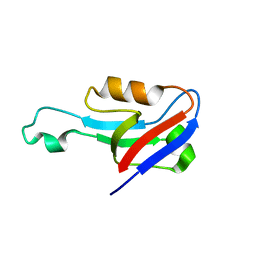 | |
1ODZ
 
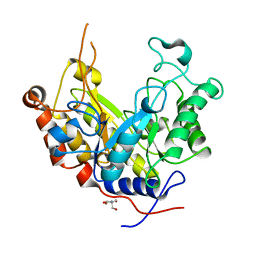 | | Expansion of the glycosynthase repertoire to produce defined manno-oligosaccharides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Mannan endo-1,4-beta-mannosidase, SODIUM ION, ... | | Authors: | Jahn, M, Stoll, D, Warren, R.A.J, Szabo, L, Singh, P, Gilbert, H.J, Ducros, V.M.A, Davies, G.J, Withers, S.G. | | Deposit date: | 2003-03-17 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expansion of the Glycosynthase Repertoire to Produce Defined Manno-Oligosaccharides
Chem.Commun.(Camb.), 12, 2003
|
|
6E3Y
 
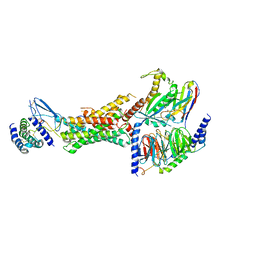 | | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | | Descriptor: | Calcitonin gene-related peptide 1, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Deganutti, G, Glukhova, A, Koole, C, Peat, T.S, Radjainia, M, Plitzko, J.M, Baumeister, W, Miller, L.J, Hay, D.L, Christopoulos, A, Reynolds, C.A, Wootten, D, Sexton, P.M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor.
Nature, 561, 2018
|
|
3VMV
 
 | | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 | | Descriptor: | Pectate lyase, SULFATE ION | | Authors: | Zheng, Y, Huang, C.H, Liu, W, Ko, T.P, Xue, Y, Zhou, C, Zhang, G, Guo, R.T, Ma, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure and substrate-binding mode of a novel pectate lyase from alkaliphilic Bacillus sp. N16-5.
Biochem.Biophys.Res.Commun., 420, 2012
|
|
6EF5
 
 | | 14-3-3 with peptide | | Descriptor: | 14-3-3 protein zeta/delta, ARG-SER-LEU-SEP-ALA-PRO-GLY, LYS-LEU-SEP-LEU-GLN | | Authors: | Dhagat, U, Langendorf, C.G, Parker, M.W.P, Oakhill, J.S, Scott, J.W. | | Deposit date: | 2018-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | 14-3-3 with peptide
To Be Published
|
|
1O2P
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-CHLORO-1H-BENZIMIDAZOL-2-YL}-6-ISOBUTOXYBENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
1O2W
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-(3-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-5-BROMO-4-OXIDOPHENYL)SUCCINATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | Deposit date: | 2003-03-06 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
5LNM
 
 | | Crystal structure of D1050E mutant of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana | | Descriptor: | Histidine kinase CKI1 | | Authors: | Otrusinova, O, Demo, G, Kaderavek, P, Jansen, S, Jasenakova, Z, Pekarova, B, Janda, L, Wimmerova, M, Hejatko, J, Zidek, L. | | Deposit date: | 2016-08-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conformational dynamics are a key factor in signaling mediated by the receiver domain of a sensor histidine kinase from Arabidopsis thaliana.
J. Biol. Chem., 292, 2017
|
|
1O34
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}PYRIDIN-3-OLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | Deposit date: | 2003-03-06 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
2ZWZ
 
 | | alpha-L-fucosidase complexed with inhibitor, Core1 | | Descriptor: | (2R,3R,4R,5R,6S)-2-(aminomethyl)-6-methylpiperidine-3,4,5-triol, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
5LNN
 
 | | Crystal structure of D1050A mutant of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana | | Descriptor: | Histidine kinase CKI1 | | Authors: | Otrusinova, O, Demo, G, Kaderavek, P, Jansen, S, Jasenakova, Z, Pekarova, B, Janda, L, Wimmerova, M, Hejatko, J, Zidek, L. | | Deposit date: | 2016-08-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational dynamics are a key factor in signaling mediated by the receiver domain of a sensor histidine kinase from Arabidopsis thaliana.
J. Biol. Chem., 292, 2017
|
|
1O3B
 
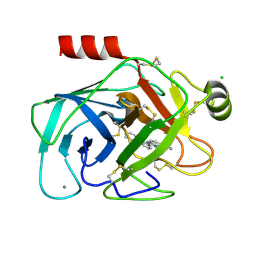 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 3-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-1,1'-BIPHENYL-2-OLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | Deposit date: | 2003-03-06 | | Release date: | 2003-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
2ZXD
 
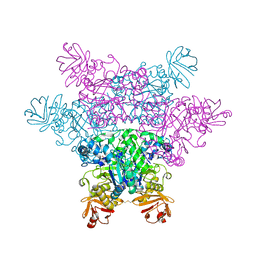 | | alpha-L-fucosidase complexed with inhibitor, iso-6FNJ | | Descriptor: | (2S,3R,4S,5R)-2-(1-methylethyl)piperidine-3,4,5-triol, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
5LNU
 
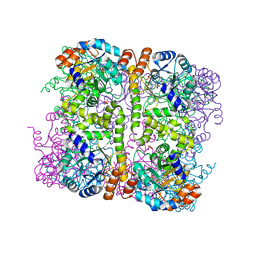 | | Crystal structure of Arabidopsis thaliana Pdx1-I320 complex | | Descriptor: | (4~{S})-4-azanyl-5-oxidanyl-pent-1-en-3-one, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
