1N9V
 
 | | Differences and Similarities in Solution Structures of Angiotensin I & II: Implication for Structure-Function Relationship. | | Descriptor: | Angiotensin II | | Authors: | Spyroulias, G.A, Nikolakopoulou, P, Tzakos, A, Gerothanassis, I.P, Magafa, V, Manessi-Zoupa, E, Cordopatis, P. | | Deposit date: | 2002-11-26 | | Release date: | 2003-07-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of angiotensin I & II. Implication for structure-function relationship.
Eur.J.Biochem., 270, 2003
|
|
2V4Y
 
 | | THE STRUCTURE OF E. COLI UMP KINASE IN COMPLEX WITH ITS ALLOSTERIC REGULATOR GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, URIDYLATE KINASE | | Authors: | Meyer, P, Evrin, C, Briozzo, P, Joly, N, Barzu, O, Gilles, A.M. | | Deposit date: | 2008-09-30 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Characterization of Escherichia Coli Ump Kinase in Complex with its Allosteric Regulator GTP.
J.Biol.Chem., 283, 2008
|
|
5F8H
 
 | | Enterovirus 71 Polymerase Elongation Complex (C1S1/2 Form) | | Descriptor: | Genome polyprotein, MAGNESIUM ION, RNA (35-MER), ... | | Authors: | Shu, B, Gong, P. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6PTA
 
 | | Crystal structure of the ARF family small GTPase ARF1 from Candida albicans in complex with GDP | | Descriptor: | ADP-ribosylation factor, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-15 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the ARF family small GTPase ARF1 from Candida albicans in complex with GDP
To Be Published
|
|
1G9M
 
 | | HIV-1 HXBC2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Kwong, P.D, Wyatt, R, Majeed, S, Robinson, J, Sweet, R.W, Sodroski, J, Hendrickson, W.A. | | Deposit date: | 2000-11-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates.
Structure Fold.Des., 8, 2000
|
|
5AJA
 
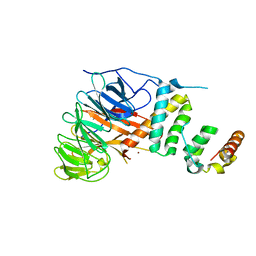 | | Crystal structure of mandrill SAMHD1 (amino acid residues 1-114) bound to Vpx isolated from mandrill and human DCAF1 (amino acid residues 1058-1396) | | Descriptor: | PROTEIN VPRBP, SAM DOMAIN AND HD DOMAIN-CONTAINING PROTEIN, VPX PROTEIN, ... | | Authors: | Schwefel, D, Boucherit, V.C, Christodoulou, E, Walker, P.A, Stoye, J.P, Bishop, K.N, Taylor, I.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Molecular Determinants for Recognition of Divergent Samhd1 Proteins by the Lentiviral Accessory Protein Vpx.
Cell Host Microbe., 17, 2015
|
|
7C9C
 
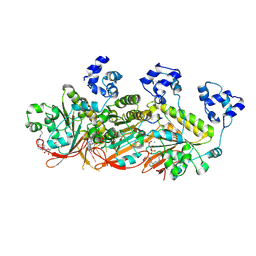 | | Human DMC1 pre-synaptic complexes | | Descriptor: | CALCIUM ION, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Meiotic recombination protein DMC1/LIM15 homolog, ... | | Authors: | Luo, S.C, Yeh, H.Y, Chi, P, Ho, M.C, Tsai, M.D. | | Deposit date: | 2020-06-05 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures.
Nat Commun, 12, 2021
|
|
6X2A
 
 | |
8BXK
 
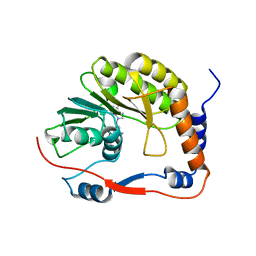 | | Ntaya virus methyltransferase | | Descriptor: | Ntaya virus methyltransferase, SINEFUNGIN | | Authors: | Boura, E, Krafcikova, P. | | Deposit date: | 2022-12-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Ntaya virus methyltransferase
To Be Published
|
|
1BLS
 
 | |
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
6ZHP
 
 | |
6WII
 
 | | Crystal structure of the K. pneumoniae LpxH/JH-LPH-41 complex | | Descriptor: | 5-({[4-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)phenyl]carbamoyl}amino)-N-hydroxypentanamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Cochrane, C.S, Cho, J, Fenton, B.A, Zhou, P. | | Deposit date: | 2020-04-09 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and evaluation of sulfonyl piperazine LpxH inhibitors.
Bioorg.Chem., 102, 2020
|
|
7V1I
 
 | |
1BK0
 
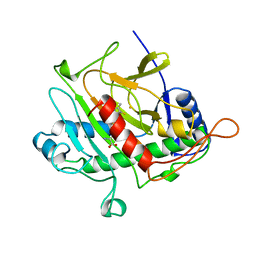 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACV-FE COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Roach, P.L, Clifton, I.J, Hensgens, C.M.H, Shibata, N, Schofield, C.J, Hajdu, J, Baldwin, J.E. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation.
Nature, 387, 1997
|
|
6PTY
 
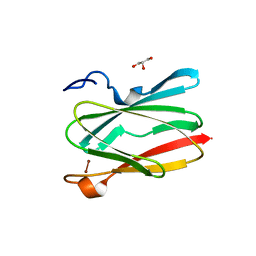 | | Soluble model of human CuA (Tt3Lh) | | Descriptor: | Cytochrome c oxidase subunit 2, DINUCLEAR COPPER ION, GLYCEROL, ... | | Authors: | Giannini, E, Lisa, M.N, Morgada, M.N, Alzari, P.M, Vila, A.J. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unexpected electron spin density on the axial methionine ligand in CuAsuggests its involvement in electron pathways.
Chem.Commun.(Camb.), 56, 2020
|
|
7K4X
 
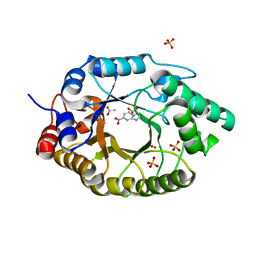 | | Crystal structure of Kemp Eliminase HG3.7 in complex with the transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
6YYK
 
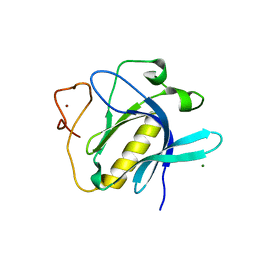 | | Crystal Structure of 1,5-dimethylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 1,5-dimethyl-3~{H}-indol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
7V1G
 
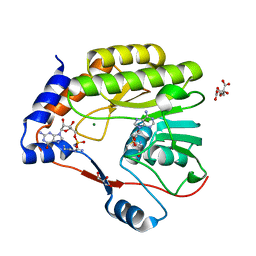 | |
7UPT
 
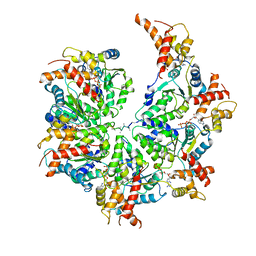 | | Human mitochondrial AAA protein ATAD1 (with a catalytic dead mutation) in complex with a peptide substrate (open conformation) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, L, Toutkoushian, H, Belyy, V, Kokontis, C, Walter, P. | | Deposit date: | 2022-04-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conserved structural elements specialize ATAD1 as a membrane protein extraction machine.
Elife, 11, 2022
|
|
7C64
 
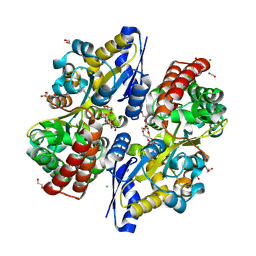 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in an open state (Form II) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CARBON DIOXIDE, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7V1E
 
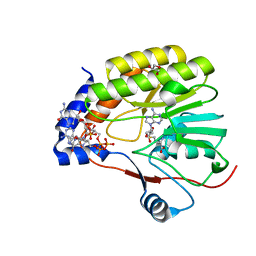 | |
7C8G
 
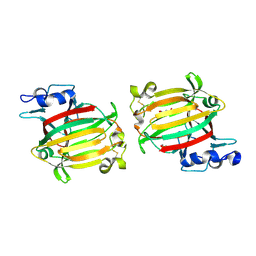 | | Structure of alginate lyase AlyC3 | | Descriptor: | Alginate lyase AlyC3, GLYCEROL, SUCCINIC ACID | | Authors: | Zhang, Y.Z, Xu, F, Chen, X.L, Wang, P. | | Deposit date: | 2020-05-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic.
J.Biol.Chem., 295, 2020
|
|
6YYF
 
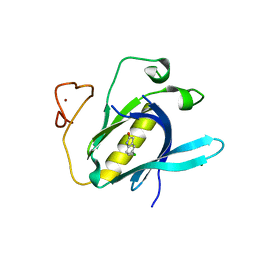 | | Crystal Structure of 5-chloroindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-chloranyl-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6WT5
 
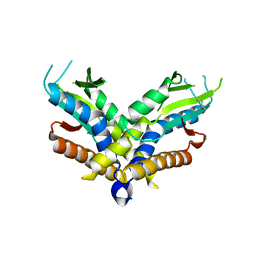 | | Structure of a bacterial STING receptor from Capnocytophaga granulosa | | Descriptor: | Bacterial STING | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
