5HPK
 
 | | System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes: NEDD4L and UbV NL.1 | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like, Ubiquitin variant NL.1 | | Authors: | Wu, K.-P, Mukherjee, M, Mercredi, P.Y, Schulman, B.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
6OKV
 
 | |
5HOH
 
 | | RIBONUCLEASE T1 (ASN9ALA/THR93ALA DOUBLEMUTANT) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
8CJ5
 
 | |
8OEA
 
 | |
2REQ
 
 | |
6I1A
 
 | | Crystal structure of rutinosidase from Aspergillus niger | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, rutinosidase | | Authors: | Pachl, P, Rezacova, P, Kapesova, J. | | Deposit date: | 2018-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rutinosidase from Aspergillus niger: crystal structure and insight into the enzymatic activity.
Febs J., 287, 2020
|
|
6OOL
 
 | | Structural elucidation of the Ectodomain of mouse UNC5H2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Netrin receptor UNC5B, ... | | Authors: | Stetefeld, J, McDougall, M.D, Loewen, P.C, Moya, A, Meier, M. | | Deposit date: | 2019-04-23 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural elucidation of the Ectodomain of mouse UNC5H2
To be published
|
|
5HN2
 
 | |
7AA2
 
 | | Chaetomium thermophilum FAD-dependent oxidoreductase in complex with ABTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7M0G
 
 | |
6XIB
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 30 | | Descriptor: | GLYCEROL, Peptide 30, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6E7P
 
 | | cryo-EM structure of human TRPML1 with PI35P2 | | Descriptor: | (1R,2S,3S,4R,5S,6R)-5-{[(R)-[(2R)-2,3-bis{[(1S)-1-hydroxyoctyl]oxy}propoxy](hydroxy)phosphoryl]oxy}-2,4,6-trihydroxycyclohexane-1,3-diyl bis[dihydrogen (phosphate)], Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
7S2H
 
 | | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation) | | Descriptor: | CITRATE ANION, Glucokinase 1, putative, ... | | Authors: | Kearns, S.P, Daneshian, L, Swartz, P.D, Carey, S.M, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation)
To Be Published
|
|
5HUM
 
 | | The crystal structure of neuraminidase from A/Sichuan/26221/2014 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, neuraminidase | | Authors: | Yang, H, Carney, P.J, Guo, Z, Chang, J.C, Stevens, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Characterizations of Surface Proteins Hemagglutinin and Neuraminidase from Recent H5Nx Avian Influenza Viruses.
J.Virol., 90, 2016
|
|
6XIC
 
 | | PCSK9(deltaCRD) in complex with cyclic peptide 40 | | Descriptor: | GLYCEROL, Peptide 40, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
5HNJ
 
 | |
8HE6
 
 | | Crystal structure of a fosfomycin and bleomycin resistant protein (ALL3014) from Anabaena/Nostoc cyanobacterium at 1.70 A resolution | | Descriptor: | All3014 protein, CALCIUM ION, MAGNESIUM ION | | Authors: | Chatterjee, A, Singh, P.K, Singh, T.P, Marina, A, Sharma, S, Rai, L.C. | | Deposit date: | 2022-11-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a fosfomycin and bleomycin resistant protein (ALL3014) from Anabaena/Nostoc cyanobacterium at 1.70 A resolution
To Be Published
|
|
2RSL
 
 | |
6ZUF
 
 | | Urea-based Foldamer Inhibitor chimera C2 in complex with ASF1 Histone chaperone | | Descriptor: | C2 foldamer/peptide hybrid inhibitor of histone chaperone ASF1, GLYCEROL, Histone chaperone ASF1A, ... | | Authors: | Bakail, M, Mbianda, J, Perrin, E.M, Guerois, R, Legrand, P, Traore, S, Douat, C, Guichard, G, Ochsenbein, F. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Optimal anchoring of a foldamer inhibitor of ASF1 histone chaperone through backbone plasticity.
Sci Adv, 7, 2021
|
|
7RWL
 
 | | Envelope-associated Adeno-associated virus serotype 2 | | Descriptor: | Capsid protein VP1 | | Authors: | Hull, J.A, Mietzsch, M, Chipman, P, Strugatsky, D, McKenna, R. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural characterization of an envelope-associated adeno-associated virus type 2 capsid.
Virology, 565, 2021
|
|
6E7Y
 
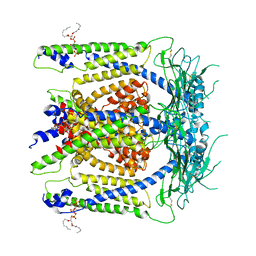 | | cryo-EM structure of human TRPML1 with PI45P2 | | Descriptor: | Mucolipin-1, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-28 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis for PtdInsP2-mediated human TRPML1 regulation.
Nat Commun, 9, 2018
|
|
1OJW
 
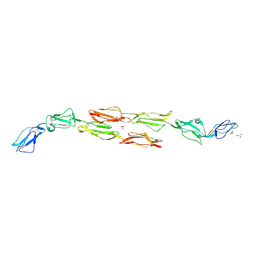 | | Decay accelerating factor (CD55): the structure of an intact human complement regulator. | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, SULFATE ION | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, C.M, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2R8G
 
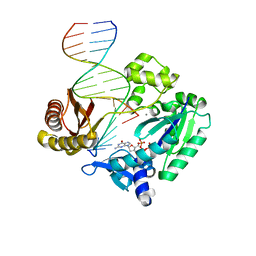 | | Selectivity of Nucleoside Triphosphate Incorporation Opposite 1,N2-Propanodeoxyguanosine (PdG) by the Sulfolobus solfataricus DNA Polymerase Dpo4 Polymerase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DGP*DGP*DGP*DGP*DGP*DAP*DAP*DGP*DGP*DAP*DTP*DTP*DT)-3', 5'-D(*DTP*DCP*DAP*DCP*(P)P*DGP*DAP*DAP*DAP*DTP*DCP*DCP*DTP*DTP*DCP*DCP*DCP*DCP*DC)-3', ... | | Authors: | Wang, Y, Saleh, S, Marnette, L.J, Egli, M, Stone, M.P. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insertion of dNTPs opposite the 1,N2-propanodeoxyguanosine adduct by Sulfolobus solfataricus P2 DNA polymerase IV
Biochemistry, 47, 2008
|
|
8BYP
 
 | |
