1TS4
 
 | | Q139K MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
5WL5
 
 | | Crystal structure of chalcone isomerase engineered from ancestral inference (ancR5) | | Descriptor: | CHLORIDE ION, Engineered Chalcone Isomerase ancR5, SULFATE ION | | Authors: | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.513 Å) | | Cite: | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
5WL7
 
 | | Crystal structure of chalcone isomerase engineered from ancestral inference (ancCHI*) | | Descriptor: | CHLORIDE ION, Engineered Chalcone Isomerase ancCHI* | | Authors: | Burke, J.R, Kaltenbach, M, Tawfik, D.S, Noel, J.P. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of chalcone isomerase from a noncatalytic ancestor.
Nat. Chem. Biol., 14, 2018
|
|
3MWA
 
 | |
3FYD
 
 | |
4K42
 
 | | Crystal structure of actin in complex with synthetic AplC tail analogue SF01 [(3R,4S,5R,6S,10R,11R,12R)-11-(acetyloxy)-1-(benzyloxy)-14-[formyl(methyl)amino]-5-hydroxy-4,6,10,12-tetramethyl-9-oxotetradecan-3-yl propanoate] | | Descriptor: | (3R,4S,5R,6S,10R,11R,12R)-11-(acetyloxy)-1-(benzyloxy)-14-[formyl(methyl)amino]-5-hydroxy-4,6,10,12-tetramethyl-9-oxotetradecan-3-yl propanoate, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Pereira, J.H, Petchprayoon, C, Moriarty, N.W, Fink, S.J, Cecere, G, Paterson, I, Adams, P.D, Marriott, G. | | Deposit date: | 2013-04-11 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical studies of actin in complex with synthetic macrolide tail analogues.
Chemmedchem, 9, 2014
|
|
8B3E
 
 | | Variant Surface Glycoprotein VSG397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Variant surface glycoprotein 397 | | Authors: | Zeelen, J.P, Stebbins, C.E, Dakovic, S, Foti, K. | | Deposit date: | 2022-09-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural similarities between the metacyclic and bloodstream form variant surface glycoproteins of the African trypanosome.
Plos Negl Trop Dis, 17, 2023
|
|
5EK8
 
 | | Crystal structure of a 9R-lipoxygenase from Cyanothece PCC8801 at 2.7 Angstroms | | Descriptor: | FE (II) ION, Lipoxygenase, SODIUM ION | | Authors: | Feussner, I, Ficner, R, Neumann, P, Newie, J, Andreou, A, Einsle, O. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a lipoxygenase from Cyanothece sp. may reveal novel features for substrate acquisition.
J.Lipid Res., 57, 2016
|
|
2K2Q
 
 | | complex structure of the external thioesterase of the Surfactin-synthetase with a carrier domain | | Descriptor: | Surfactin synthetase thioesterase subunit, Tyrocidine synthetase 3 (Tyrocidine synthetase III) | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guntert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Dotsch, V. | | Deposit date: | 2008-04-10 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase.
Nature, 454, 2008
|
|
2X2P
 
 | | The flavoprotein NrdI from Bacillus cereus with the initially semiquinone FMN cofactor in an intermediate radiation reduced state | | Descriptor: | CACODYLATE ION, FLAVIN MONONUCLEOTIDE, NRDI PROTEIN, ... | | Authors: | Rohr, A.K, Hersleth, H.P, Andersson, K.K. | | Deposit date: | 2010-01-15 | | Release date: | 2010-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Tracking Flavin Conformations in Protein Crystal Structures with Raman Spectroscopy and Qm/Mm Calculations
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
5EOD
 
 | | Human Plasma Coagulation FXI with peptide LP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XI, LP2 | | Authors: | Wong, S.S, Ostergaard, S, Hall, G, Li, C, Williams, P.M, Stennicke, H, Emsley, J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A novel DFP tripeptide motif interacts with the coagulation factor XI apple 2 domain.
Blood, 127, 2016
|
|
2WHQ
 
 | | Crystal structure of acetylcholinesterase, phosphonylated by sarin (aged) in complex with HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.G, Schneider, G, Pang, Y.P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
4KO9
 
 | |
1D0E
 
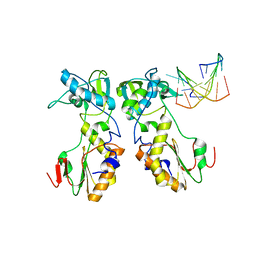 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | 5'-D(*TP*TP*TP*CP*AP*TP*GP*CP*AP*TP*G)-3', REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M.L, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-09-09 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain
J.Mol.Biol., 296, 2000
|
|
2WIJ
 
 | | NONAGED FORM OF HUMAN BUTYRYLCHOLINESTERASE INHIBITED BY TABUN ANALOGUE TA5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Carletti, E, Aurbek, N, Gillon, E, Loiodice, M, Nicolet, Y, Fontecilla, J, Masson, P, Thiermann, H, Nachon, F, Worek, F. | | Deposit date: | 2009-05-12 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity analysis of aging and reactivation of human butyrylcholinesterase inhibited by analogues of tabun.
Biochem. J., 421, 2009
|
|
6HHK
 
 | |
1KGP
 
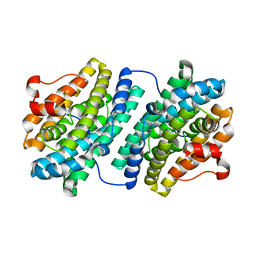 | | R2F from Corynebacterium Ammoniagenes in its Mn substituted form | | Descriptor: | MANGANESE (II) ION, Ribonucleotide reductase protein R2F | | Authors: | Hogbom, M, Huque, Y, Sjoberg, B.M, Nordlund, P. | | Deposit date: | 2001-11-28 | | Release date: | 2001-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the di-iron/radical protein of ribonucleotide reductase from Corynebacterium ammoniagenes.
Biochemistry, 41, 2002
|
|
6HI5
 
 | | The ATAD2 bromodomain in complex with compound 8 | | Descriptor: | (2~{R})-2-azanyl-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-1,3-thiazol-2-yl]propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6C7W
 
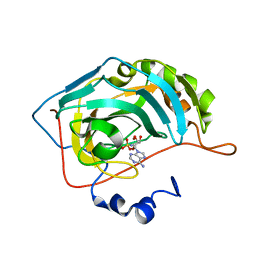 | | Carbonic anhydrase 2 in complex with [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDRO-2-FURANYL]METHYL SULFAMATE inhibitor | | Descriptor: | Carbonic anhydrase 2, SODIUM ION, ZINC ION, ... | | Authors: | Peat, T.S, Mujumdar, P, Poulsen, S.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Synthesis, structure and bioactivity of primary sulfamate-containing natural products.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
3MTC
 
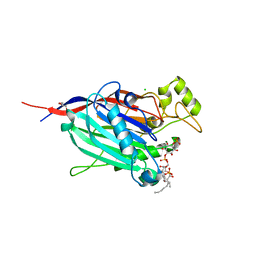 | | Crystal Structure of INPP5B in complex with phosphatidylinositol 4-phosphate | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases
Structure, 22, 2014
|
|
6HIA
 
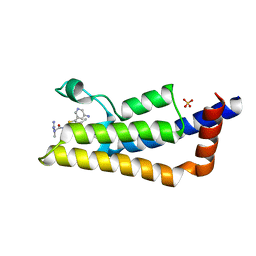 | | The ATAD2 bromodomain in complex with compound 13 | | Descriptor: | (2~{R})-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-1,3-thiazol-2-yl]-2-carbamimidamido-propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
2JLL
 
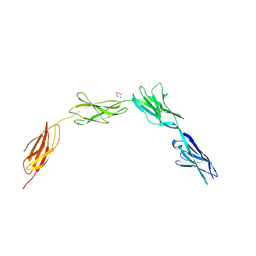 | | Crystal structure of NCAM2 IgIV-FN3II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Kulahin, N, Rasmussen, K, Kristensen, O, Kastrup, J, Berezin, V, Bock, E, Walmod, P, Gajhede, M. | | Deposit date: | 2008-09-10 | | Release date: | 2009-11-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
4KQX
 
 | | Mutant Slackia exigua KARI DDV in complex with NAD and an inhibitor | | Descriptor: | HISTIDINE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Brinkmann-Chen, S, Flock, T, Cahn, J.K.B, Snow, C.D, Brustad, E.M, Mcintosh, J.A, Meinhold, P, Zhang, L, Arnold, F.H. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5WTU
 
 | |
3RHB
 
 | | Crystal structure of the apo form of glutaredoxin C5 from Arabidopsis thaliana | | Descriptor: | GLUTATHIONE, Glutaredoxin-C5, chloroplastic, ... | | Authors: | Roret, T, Couturier, J, Tsan, P, Jacquot, J.P, Rouhier, N, Didierjean, C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Arabidopsis chloroplastic glutaredoxin c5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins.
J.Biol.Chem., 286, 2011
|
|
