1N1P
 
 | |
6LZ7
 
 | |
6LOT
 
 | | Crystal structure of DUSP22 mutant_N128D | | 分子名称: | Dual specificity protein phosphatase 22, SULFATE ION | | 著者 | Lai, C.H, Lyu, P.C. | | 登録日 | 2020-01-07 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural Insights into the Active Site Formation of DUSP22 in N-loop-containing Protein Tyrosine Phosphatases.
Int J Mol Sci, 21, 2020
|
|
3LKO
 
 | | Crystal Structure of HLA B*3501 in complex with influenza NP418 epitope from 1934 strain | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | 著者 | Gras, S, Kedzierski, L, Valkenburg, S.A, Liu, Y.C, Denholm, J, Richards, M, Rimmelzwaan, G.F, Doherty, P.C, Turner, S.J, Rossjohn, J, Kedzierska, K. | | 登録日 | 2010-01-27 | | 公開日 | 2010-07-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4N6R
 
 | | Crystal structure of VosA-VelB-complex | | 分子名称: | SULFATE ION, VelB, VosA | | 著者 | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | 登録日 | 2013-10-14 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
6D33
 
 | | Crystal structure of BH1352 2-deoxyribose-5-phosphate from Bacillus halodurans | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyribose-phosphate aldolase, GLYCEROL | | 著者 | Stogios, P.J, Skarina, T, Kim, T, Yim, V, Yakunin, A, Savchenko, A. | | 登録日 | 2018-04-14 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Rational engineering of 2-deoxyribose-5-phosphate aldolases for the biosynthesis of (R)-1,3-butanediol.
J.Biol.Chem., 295, 2020
|
|
1MUO
 
 | | CRYSTAL STRUCTURE OF AURORA-2, AN ONCOGENIC SERINE-THREONINE KINASE | | 分子名称: | ADENOSINE, Aurora-related kinase 1 | | 著者 | Cheetham, G.M.T, Knegtel, R.M.A, Coll, J.T, Renwick, S.B, Swenson, L, Weber, P, Lippke, J.A, Austen, D.A. | | 登録日 | 2002-09-24 | | 公開日 | 2003-04-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of Aurora-2, an Oncogenic Serine/Threonine Kinase
J.Biol.Chem., 277, 2002
|
|
3LM0
 
 | | Crystal Structure of human Serine/Threonine Kinase 17B (STK17B) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Serine/threonine-protein kinase 17B, ... | | 著者 | Ugochukwu, E, Soundararajan, M, Rellos, P, Fedorov, O, Phillips, C, Wang, J, Hapka, E, Filippakopoulos, P, Chaikuad, A, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2010-01-29 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 |
|
|
1MZ6
 
 | | Trypanosoma rangeli sialidase in complex with the inhibitor DANA | | 分子名称: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, sialidase | | 著者 | Buschiazzo, A, Tavares, G.A, Campetella, O, Spinelli, S, Cremona, M.L, Paris, G, Amaya, M.F, Frasch, A.C.C, Alzari, P.M. | | 登録日 | 2002-10-05 | | 公開日 | 2002-10-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of sialyltransferase activity in trypanosomal sialidases
Embo J., 19, 2000
|
|
3LDP
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with small molecule inhibitor | | 分子名称: | 78 kDa glucose-regulated protein, 8-[(quinolin-2-ylmethyl)amino]adenosine | | 著者 | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | 登録日 | 2010-01-13 | | 公開日 | 2011-01-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
4MO4
 
 | | Crystal structure of AnmK bound to AMPPCP | | 分子名称: | Anhydro-N-acetylmuramic acid kinase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | 著者 | Bacik, J.P, Mark, B.L. | | 登録日 | 2013-09-11 | | 公開日 | 2014-01-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Conformational Itinerary of Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase during Its Catalytic Cycle.
J.Biol.Chem., 289, 2014
|
|
6D4P
 
 | | Ube2D1 in complex with ubiquitin variant Ubv.D1.1 | | 分子名称: | Ubiquitin Variant Ubv.D1.1, Ubiquitin-conjugating enzyme E2 D1 | | 著者 | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | 登録日 | 2018-04-18 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
3IXE
 
 | | Structural basis of competition between PINCH1 and PINCH2 for binding to the ankyrin repeat domain of integrin-linked kinase | | 分子名称: | Integrin-linked protein kinase, LIM and senescent cell antigen-like-containing domain protein 2, ZINC ION | | 著者 | Chiswell, B.P, Stiegler, A.L, Boggon, T.J, Calderwood, D.A. | | 登録日 | 2009-09-03 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of competition between PINCH1 and PINCH2 for binding to the ankyrin repeat domain of integrin-linked kinase.
J.Struct.Biol., 170, 2010
|
|
1MRV
 
 | | crystal structure of an inactive Akt2 kinase domain | | 分子名称: | RAC-beta serine/threonine kinase | | 著者 | Huang, X, Begley, M, Morgenstern, K.A, Gu, Y, Rose, P, Zhao, H, Zhu, X. | | 登録日 | 2002-09-18 | | 公開日 | 2003-09-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of an inactive akt2 kinase domain
Structure, 11, 2003
|
|
3IY2
 
 | | Variable domains of the computer generated model (WAM) of Fab 6 fitted into the cryoEM reconstruction of the virus-Fab 6 complex | | 分子名称: | Antibody 6, heavy chain, light chain | | 著者 | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | 登録日 | 2009-04-09 | | 公開日 | 2009-05-12 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
1MRE
 
 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | 著者 | Pokkuluri, P.R, Cygler, M. | | 登録日 | 1994-06-13 | | 公開日 | 1995-02-14 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
4N9W
 
 | | Crystal structure of phosphatidyl mannosyltransferase PimA | | 分子名称: | 1,2-ETHANEDIOL, GDP-mannose-dependent alpha-(1-2)-phosphatidylinositol mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Giganti, D, Albesa-Jove, D, Bellinzoni, M, Guerin, M.E, Alzari, P.M. | | 登録日 | 2013-10-21 | | 公開日 | 2014-11-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Secondary structure reshuffling modulates glycosyltransferase function at the membrane.
Nat.Chem.Biol., 11, 2015
|
|
1MS3
 
 | | Monoclinic form of Trypanosoma cruzi trans-sialidase | | 分子名称: | trans-sialidase | | 著者 | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | 登録日 | 2002-09-19 | | 公開日 | 2003-03-25 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MT3
 
 | | Crystal Structure of the Tricorn Interacting Factor Selenomethionine-F1 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proline iminopeptidase | | 著者 | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | 登録日 | 2002-09-20 | | 公開日 | 2002-11-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
4RC6
 
 | | Crystal structure of cyanobacterial aldehyde-deformylating oxygenase 122F mutant | | 分子名称: | Aldehyde decarbonylase, FE (II) ION | | 著者 | Jia, C.J, Li, M, Li, J.J, Zhang, J.J, Zhang, H.M, Cao, P, Pan, X.W, Lu, X.F, Chang, W.R. | | 登録日 | 2014-09-14 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of aldehyde-deformylating oxygenases.
Protein Cell, 6, 2015
|
|
4N0K
 
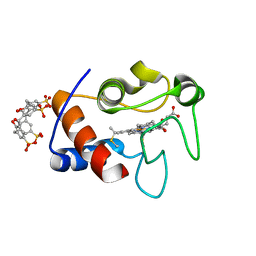 | | Atomic resolution crystal structure of a cytochrome c-calixarene complex | | 分子名称: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, Cytochrome c iso-1, HEME C | | 著者 | McGovern, R.E, Pye, V.E, Crowley, P.B. | | 登録日 | 2013-10-02 | | 公開日 | 2014-09-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | A cytochrome c-calixarene structure at atomic resolution
To be Published
|
|
6DC7
 
 | |
4N1B
 
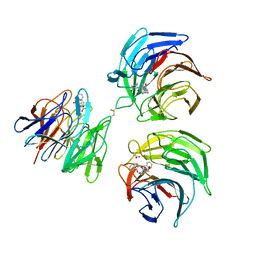 | | STRUCTURE OF KEAP1 KELCH DOMAIN WITH(1S,2R)-2-[(1S)-1-[(1-oxo-2,3-dihydro-1H-isoindol-2-Yl)methyl]-1,2,3,4-tetrahydroisoquinoline-2-Carbonyl]cyclohexane-1-carboxylic acid | | 分子名称: | (1S,2R)-2-{[(1S)-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | 著者 | Smith, M.A, Duclos, S, Beaumont, E, Kwong, J, Brooks, M, Barker, J, Jnoff, E, Brookfield, F, Courade, J.P, Barker, O, Fryatt, T, Albrecht, C, Bromidge, S. | | 登録日 | 2013-10-03 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
4MQY
 
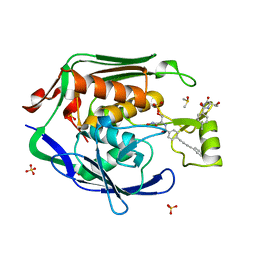 | | Crystal Structure of the Escherichia coli LpxC/LPC-138 complex | | 分子名称: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-2-methyl-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Najeeb, J, Zhou, P. | | 登録日 | 2013-09-17 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.005 Å) | | 主引用文献 | Structural Basis of the Promiscuous Inhibitor Susceptibility of Escherichia coli LpxC.
Acs Chem.Biol., 9, 2014
|
|
3IXX
 
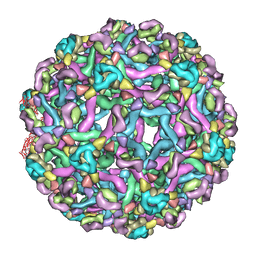 | | The pseudo-atomic structure of West Nile immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | 分子名称: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | 著者 | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | 登録日 | 2009-02-26 | | 公開日 | 2009-10-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (15 Å) | | 主引用文献 | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
