1HEO
 
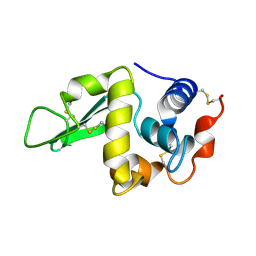 | |
6Y9X
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,7) | | 分子名称: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2020-03-10 | | 公開日 | 2020-08-19 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7OU0
 
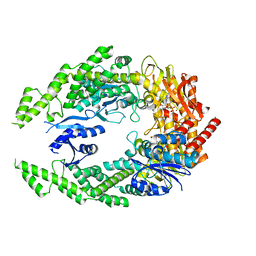 | | The structure of MutS bound to two molecules of ADP-Vanadate | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS, MAGNESIUM ION, ... | | 著者 | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | 登録日 | 2021-06-10 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3MOK
 
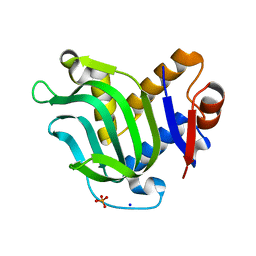 | | Structure of Apo HasAp from Pseudomonas aeruginosa to 1.55A Resolution | | 分子名称: | Heme acquisition protein HasAp, PHOSPHATE ION, SODIUM ION | | 著者 | Lovell, S, Battaile, K.P, Jepkorir, G, Rodriguez, J.C, Rui, H, Im, W, Alontaga, A.Y, Yukl, E, Moenne-Loccoz, P, Rivera, M. | | 登録日 | 2010-04-22 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural, NMR Spectroscopic, and Computational Investigation of Hemin Loading in the Hemophore HasAp from Pseudomonas aeruginosa.
J.Am.Chem.Soc., 132, 2010
|
|
6BN9
 
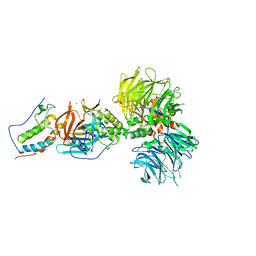 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET70 PROTAC | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (4.382 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
7R7E
 
 | |
7OW8
 
 | | CryoEM structure of the ABC transporter BmrA E504A mutant in complex with ATP-Mg | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | 著者 | Gobet, A, Schoehn, G, Falson, P, Chaptal, V. | | 登録日 | 2021-06-17 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
1LAI
 
 | | Solution Structure of the B-DNA Duplex CGCGGTGTCCGCG. | | 分子名称: | 5'-D(*CP*GP*CP*GP*GP*AP*CP*AP*CP*CP*GP*CP*G)-3', 5'-D(*CP*GP*CP*GP*GP*TP*GP*TP*CP*CP*GP*CP*G)-3' | | 著者 | Weisenseel, J.P, Reddy, G.R, Marnett, L.J, Stone, M.P. | | 登録日 | 2002-03-28 | | 公開日 | 2002-04-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of an oligodeoxynucleotide containing a 1,N(2)-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2.
Chem.Res.Toxicol., 15, 2002
|
|
6BR8
 
 | | Structure of A6 reveals a novel lipid transporter | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Protein A6 homolog, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | 著者 | Deng, J, Peng, S, Pathak, P. | | 登録日 | 2017-11-30 | | 公開日 | 2018-06-20 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of a lipid-bound viral membrane assembly protein reveals a modality for enclosing the lipid bilayer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7R36
 
 | | Difference-refined structure of fatty acid photodecarboxylase 2 microsecond following 400-nm laser irradiation of the dark-state determined by SFX | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | 著者 | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | 登録日 | 2022-02-06 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1D6F
 
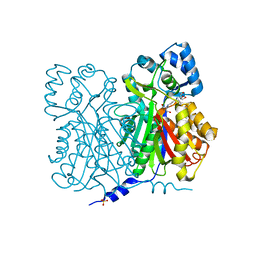 | | CHALCONE SYNTHASE C164A MUTANT | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHALCONE SYNTHASE, SULFATE ION | | 著者 | Jez, J.M, Ferrer, J.L, Bowman, M.E, Dixon, R.A, Noel, J.P. | | 登録日 | 1999-10-13 | | 公開日 | 2000-02-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase.
Biochemistry, 39, 2000
|
|
7R33
 
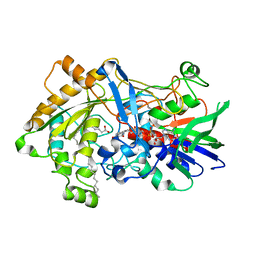 | | Difference-refined structure of fatty acid photodecarboxylase 20 ps following 400-nm laser irradiation of the dark-state determined by SFX | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | 著者 | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | 登録日 | 2022-02-06 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3MMA
 
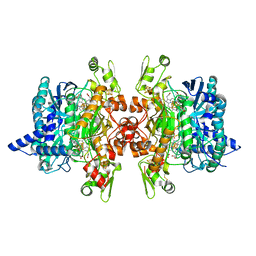 | | Dissimilatory sulfite reductase phosphate complex | | 分子名称: | IRON/SULFUR CLUSTER, PHOSPHATE ION, SIROHEME, ... | | 著者 | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | 登録日 | 2010-04-19 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
7R34
 
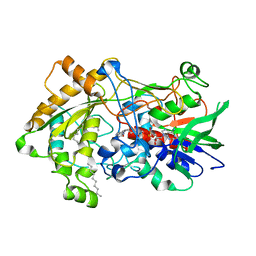 | | Difference-refined structure of fatty acid photodecarboxylase 900 ps following 400-nm laser irradiation of the dark-state determined by SFX | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | 著者 | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, M. | | 登録日 | 2022-02-06 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R35
 
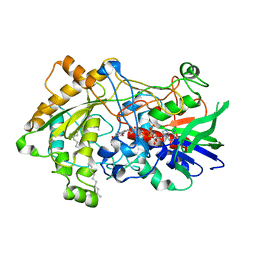 | | Difference-refined structure of fatty acid photodecarboxylase 300 ns following 400-nm laser irradiation of the dark-state determined by SFX | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | 著者 | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, De Zitter, E, Schlichting, I, Colletier, J.P, Weik, W. | | 登録日 | 2022-02-06 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Time-resolved serial femtosecond crystallography on fatty-acid photodecarboxylase: lessons learned.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6HBL
 
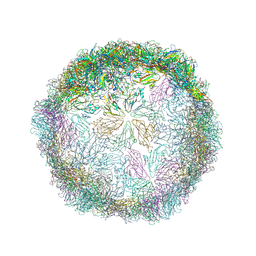 | | Echovirus 18 Open particle without three pentamers | | 分子名称: | Echovirus 18 capsid protein 1, Echovirus 18 capsid protein 2, Echovirus 18 capsid protein 3 | | 著者 | Buchta, D, Fuzik, T, Hrebik, D, Levdansky, Y, Moravcova, J, Plevka, P. | | 登録日 | 2018-08-10 | | 公開日 | 2019-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Enterovirus particles expel capsid pentamers to enable genome release.
Nat Commun, 10, 2019
|
|
6H93
 
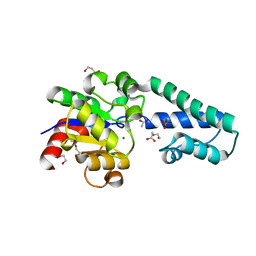 | | Beta-phosphoglucomutase from Lactococcus lactis with inorganic phosphate bound in an open conformer to 1.8 A. | | 分子名称: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ACETATE ION, ... | | 著者 | Robertson, A.J, Bisson, C, Waltho, J.P. | | 登録日 | 2018-08-03 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
7W9P
 
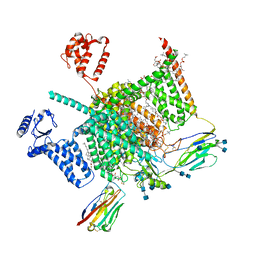 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV pi helix conformer) | | 分子名称: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | 登録日 | 2021-12-10 | | 公開日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
5EHZ
 
 | | mAChE-syn TZ2PA5 complex from an equimolar mixture of the syn/anti isomers | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-phenyl-5-[5-[3-[2-(1,2,3,4-tetrahydroacridin-9-ylamino)ethyl]-1,2,3-triazol-4-yl]pentyl]phenanthridin-5-ium-3,8-diamine, Acetylcholinesterase, ... | | 著者 | Bourne, Y, Marchot, P. | | 登録日 | 2015-10-29 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
1URL
 
 | | N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) IN COMPLEX WITH GLYCOPEPTIDE | | 分子名称: | ALA-GLY-HIS-THR-TRP-GLY-HIA, N-acetyl-alpha-neuraminic acid, SIALOADHESIN | | 著者 | Bukrinsky, J.T, Hilaire, P.M.S, Meldal, M, Crocker, P.R, Henriksen, A. | | 登録日 | 2003-10-31 | | 公開日 | 2004-10-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Complex of Sialoadhesin with a Glycopeptide Ligand
Biochim.Biophys.Acta, 1702, 2004
|
|
6YDD
 
 | | X-ray structure of LPMO. | | 分子名称: | COPPER (II) ION, LPMO lytic polysaccharide monooxygenase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | 著者 | Tandrup, T, Tryfona, T, Frandsen, K.E.H, Johansen, K.S, Dupree, P, Lo Leggio, L. | | 登録日 | 2020-03-20 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Oligosaccharide Binding and Thermostability of Two Related AA9 Lytic Polysaccharide Monooxygenases.
Biochemistry, 59, 2020
|
|
7RGY
 
 | |
4X5T
 
 | | alpha 1 glycine receptor transmembrane structure fused to the extracellular domain of GLIC | | 分子名称: | ACETATE ION, CHLORIDE ION, NICKEL (II) ION, ... | | 著者 | Sauguet, L, Corringer, P.J, Huon, C, Delarue, M. | | 登録日 | 2014-12-05 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Allosteric and hyperekplexic mutant phenotypes investigated on an alpha 1 glycine receptor transmembrane structure.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7D4O
 
 | | cyclic trinucleotide synthase CdnD in complex with ATP and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase, ... | | 著者 | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | 登録日 | 2020-09-24 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
1H61
 
 | | Structure of Pentaerythritol Tetranitrate Reductase in complex with prednisone | | 分子名称: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | 著者 | Barna, T.M, Moody, P.C.E. | | 登録日 | 2001-06-04 | | 公開日 | 2001-07-05 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
