1UGY
 
 | | Crystal structure of jacalin- mellibiose (Gal-alpha(1-6)-Glc) complex | | 分子名称: | Agglutinin alpha chain, Agglutinin beta-3 chain, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose, ... | | 著者 | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | 登録日 | 2003-06-23 | | 公開日 | 2003-09-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
7A0L
 
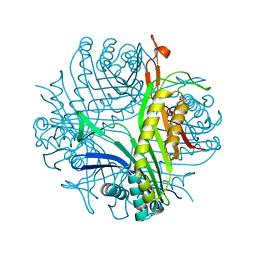 | | Joint neutron/X-ray room temperature structure of perdeuterated Aspergillus flavus urate oxidase in complex with the 8-azaxanthine inhibitor and catalytic water bound in the peroxo hole | | 分子名称: | 8-AZAXANTHINE, SODIUM ION, Uricase | | 著者 | McGregor, L, Bui, S, Blakeley, M.P, Steiner, R.A. | | 登録日 | 2020-08-09 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | NEUTRON DIFFRACTION (1.33 Å), X-RAY DIFFRACTION | | 主引用文献 | Joint neutron/X-ray crystal structure of a mechanistically relevant complex of perdeuterated urate oxidase and simulations provide insight into the hydration step of catalysis.
Iucrj, 8, 2021
|
|
2XE7
 
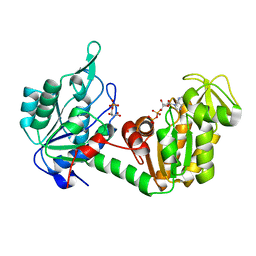 | | The complete reaction cycle of human phosphoglycerate kinase: The open ternary complex with 3PG and ADP | | 分子名称: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, PHOSPHOGLYCERATE KINASE 1 | | 著者 | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | 登録日 | 2010-05-11 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
7K5G
 
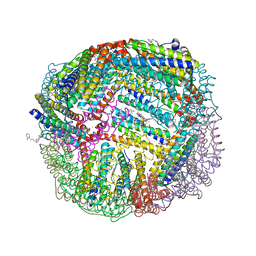 | | 1.95 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-28 | | 分子名称: | 4-{[3-(2-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | 著者 | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
4IF4
 
 | |
4K1S
 
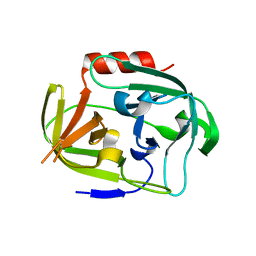 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.96 A resolution | | 分子名称: | Serine protease SplB | | 著者 | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | 登録日 | 2013-04-05 | | 公開日 | 2014-04-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
2AF6
 
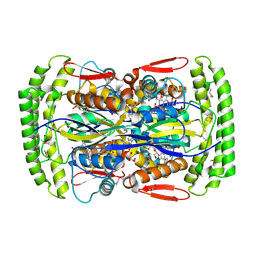 | | Crystal structure of Mycobacterium tuberculosis Flavin dependent thymidylate synthase (Mtb ThyX) in the presence of co-factor FAD and substrate analog 5-Bromo-2'-Deoxyuridine-5'-Monophosphate (BrdUMP) | | 分子名称: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Sampathkumar, P, Turley, S, Ulmer, J.E, Rhie, H.G, Sibley, C.H, Hol, W.G. | | 登録日 | 2005-07-25 | | 公開日 | 2005-10-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure of the Mycobacterium tuberculosis Flavin Dependent Thymidylate Synthase (MtbThyX) at 2.0A Resolution.
J.Mol.Biol., 352, 2005
|
|
7K4R
 
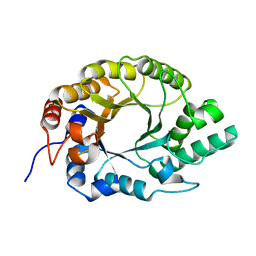 | | Crystal structure of Kemp Eliminase HG3 K50Q | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
5I9L
 
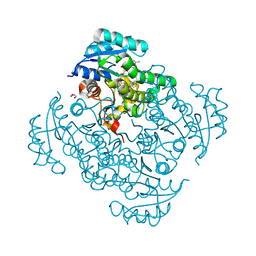 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT404 | | 分子名称: | 2-(2-chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | 著者 | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | 登録日 | 2016-02-20 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
1TG1
 
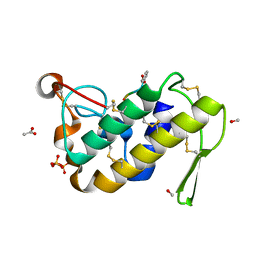 | | Crystal Structure of the complex formed between russells viper phospholipase A2 and a designed peptide inhibitor PHQ-Leu-Val-Arg-Tyr at 1.2A resolution | | 分子名称: | ACETIC ACID, METHANOL, Phospholipase A2, ... | | 著者 | Singh, N, Kaur, P, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Singh, T.P. | | 登録日 | 2004-05-28 | | 公開日 | 2004-06-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal Structure of the complex formed between russells viper phospholipase A2 and a designed peptide inhibitor Cbz-dehydro-Leu-Val-Arg-Tyr at 1.2A resolution
To be Published
|
|
7K4T
 
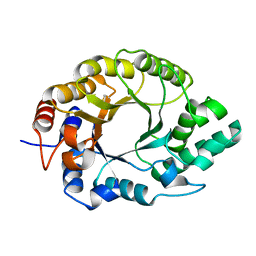 | | Crystal structure of Kemp Eliminase HG3.17 | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.999 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
3JS1
 
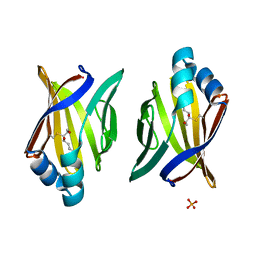 | | Crystal structure of adipocyte fatty acid binding protein covalently modified with 4-hydroxy-2-nonenal | | 分子名称: | Adipocyte fatty acid-binding protein, PHOSPHATE ION | | 著者 | Hellberg, K, Grimsrud, P.A, Kruse, A.C, Banaszak, L.J, Ohlendorf, D.H, Bernlohr, D.A. | | 登録日 | 2009-09-09 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal.
Protein Sci., 19, 2010
|
|
7KDK
 
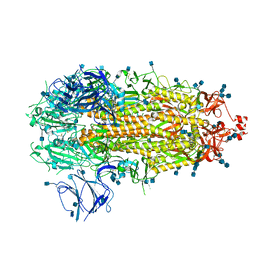 | |
6ZVM
 
 | | Botulinum neurotoxin B2 binding domain in complex with GD1a | | 分子名称: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin | | 著者 | Davies, J.R, Masuyer, G, Stenmark, P. | | 登録日 | 2020-07-25 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and Biochemical Characterization of Botulinum Neurotoxin Subtype B2 Binding to Its Receptors.
Toxins, 12, 2020
|
|
2JZW
 
 | | How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription | | 分子名称: | DNA (5'-D(*DGP*DTP*DCP*DCP*DCP*DTP*DGP*DTP*DTP*DCP*DGP*DGP*DGP*DC)-3'), HIV-1 nucleocapsid protein NCp7(12-55), ZINC ION | | 著者 | Bourbigot, S, Ramalanjaona, N, Salgado, G.F.J, Mely, Y, Roques, B.P, Bouaziz, S, Morellet, N. | | 登録日 | 2008-01-21 | | 公開日 | 2009-01-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription.
J.Mol.Biol., 383, 2008
|
|
1MR0
 
 | | SOLUTION NMR STRUCTURE OF AGRP(87-120; C105A) | | 分子名称: | AGOUTI RELATED PROTEIN | | 著者 | Jackson, P.J, Mcnulty, J.C, Yang, Y.K, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Millhauser, G.M. | | 登録日 | 2002-09-17 | | 公開日 | 2002-10-02 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity.
Biochemistry, 41, 2002
|
|
2NXZ
 
 | | HIV-1 gp120 Envelope Glycoprotein (T257S, S334A, S375W) Complexed with CD4 and Antibody 17b | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, ... | | 著者 | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | 登録日 | 2006-11-20 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
2XSO
 
 | |
1XF6
 
 | | High resolution crystal structure of phycoerythrin 545 from the marine cryptophyte rhodomonas CS24 | | 分子名称: | 15,16-DIHYDROBILIVERDIN, B-phycoerythrin beta chain, CHLORIDE ION, ... | | 著者 | Doust, A.B, Marai, C.N.J, Harrop, S.J, Wilk, K.E, Curmi, P.M.G, Scholes, G.D. | | 登録日 | 2004-09-14 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Developing a structure-function model for the cryptophyte phycoerythrin 545 using ultrahigh resolution crystallography and ultrafast laser spectroscopy
J.Mol.Biol., 344, 2004
|
|
7KE6
 
 | |
1XG0
 
 | | High resolution crystal structure of phycoerythrin 545 from the marine cryptophyte rhodomonas CS24 | | 分子名称: | 15,16-DIHYDROBILIVERDIN, B-phycoerythrin beta chain, CHLORIDE ION, ... | | 著者 | Doust, A.B, Marai, C.N.J, Harrop, S.J, Wilk, K.E, Curmi, P.M.G, Scholes, G.D. | | 登録日 | 2004-09-16 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | Developing a structure-function model for the cryptophyte phycoerythrin 545 using ultrahigh resolution crystallography and ultrafast laser spectroscopy
J.Mol.Biol., 344, 2004
|
|
7KE7
 
 | |
204D
 
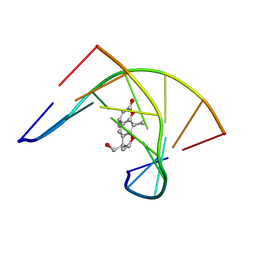 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | 分子名称: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | 著者 | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | 登録日 | 1995-04-06 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
203D
 
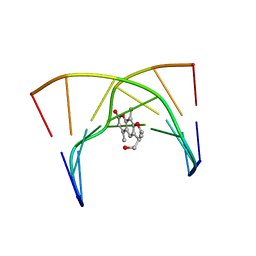 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | 分子名称: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | 著者 | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | 登録日 | 1995-04-06 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
1TJA
 
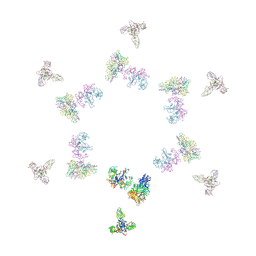 | | Fitting of gp8, gp9, and gp11 into the cryo-EM reconstruction of the bacteriophage T4 contracted tail | | 分子名称: | Baseplate structural protein Gp11, Baseplate structural protein Gp8, Baseplate structural protein Gp9 | | 著者 | Leiman, P.G, Chipman, P.R, Kostyuchenko, V.A, Mesyanzhinov, V.V, Rossmann, M.G. | | 登録日 | 2004-06-03 | | 公開日 | 2004-08-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (16 Å) | | 主引用文献 | Three-dimensional rearrangement of proteins in the tail of bacteriophage t4 on infection of its host
Cell(Cambridge,Mass.), 118, 2004
|
|
