8G55
 
 | |
7FT5
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with POB0093 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methoxyphenyl)oxane-4-carboxylic acid, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSO
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z198195770 | | Descriptor: | 1,2-ETHANEDIOL, D-GLUTAMIC ACID, GLYCINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSQ
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z1429867185 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
7FSS
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z839706072 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-bromanylpyrazol-1-yl)-~{N}-cyclopropyl-~{N}-methyl-ethanamide, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
5AZJ
 
 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with 4NP (with disulfide bridge) | | Descriptor: | 4-NITROPHENYL PHOSPHATE, GLYCEROL, Glycerol kinase | | Authors: | Balogun, E.O, Inaoka, D.K, Shiba, T, Tokuoka, S.M, Tokumasu, F, Sakamoto, K, Michels, P.A.M, Harada, S, Kita, K. | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Glycerol kinase of African trypanosomes possesses an intrinsic phosphatase activity.
Biochim Biophys Acta Gen Subj, 1861, 2017
|
|
7FSZ
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z3006151474 | | Descriptor: | (5S)-5-(difluoromethoxy)pyridin-2(5H)-one, 1,2-ETHANEDIOL, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
8G54
 
 | |
7FT4
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z48847594 | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, D-GLUTAMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
3ZLG
 
 | | Structure of group A Streptococcal enolase K362A mutant | | Descriptor: | ENOLASE, PHOSPHATE ION | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
7FT3
 
 | | SDCBP PanDDA analysis group deposition -- The PDZ domans of SDCBP in complex with Z291279160 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(trifluoromethyloxy)phenyl]thiourea, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-24 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SDCBP PanDDA analysis group deposition
To Be Published
|
|
1TLB
 
 | | Yeast coproporphyrinogen oxidase | | Descriptor: | Coproporphyrinogen III oxidase, SULFATE ION | | Authors: | Phillip, J.D, Whitby, F.G, Warby, C.A, Labbe, P, Yang, C, Pflugrath, J.W, Ferrara, J.D, Robinson, H, Kushner, J.P, Hill, C.P. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the oxygen-dependent coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
1XF2
 
 | | Structure of Fab DNA-1 complexed with dT3 | | Descriptor: | 5'-D(*TP*TP*T)-3', SULFATE ION, antibody heavy chain Fab, ... | | Authors: | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2004-09-13 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
1JB7
 
 | |
4IRI
 
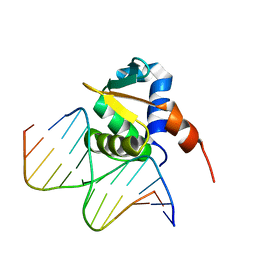 | | Auto-inhibited ERG Ets Domain-DNA Complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*G)-3'), Transcriptional regulator ERG | | Authors: | Regan, M.C, Horanyi, P.S, Pryor, E.E, Sarver, J.L, Cafiso, D.S, Bushweller, J.H. | | Deposit date: | 2013-01-14 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural and dynamic studies of the transcription factor ERG reveal DNA binding is allosterically autoinhibited.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZSZ
 
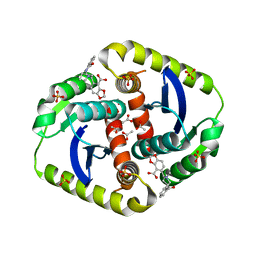 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (R)-[2-[[(2S)-BUTAN-2-YL]CARBAMOYL]PHENYL]METHYL-[(4-CARBOXY-1,3-BENZODIOXOL-5-YL)METHYL]-METHYL-AZANIUM, ACETATE ION, CHLORIDE ION, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
4L0U
 
 | | Crystal structure of Plasmodium vivax Prx1a | | Descriptor: | 2-Cys peroxiredoxin, putative, ACETATE ION | | Authors: | Gretes, M.C, Karplus, P.A. | | Deposit date: | 2013-06-01 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Observed octameric assembly of a Plasmodium yoelii peroxiredoxin can be explained by the replacement of native "ball-and-socket" interacting residues by an affinity tag.
Protein Sci., 22, 2013
|
|
6R3A
 
 | | BACTERIOPHAGE SPP1 MATURE CAPSID PROTEIN | | Descriptor: | Major capsid protein | | Authors: | Ignatiou, A, El Sadek Fadel, M, Buerger, J, Mielke, T, Topf, M, Tavares, P. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural transitions during the scaffolding-driven assembly of a viral capsid.
Nat Commun, 10, 2019
|
|
1MRR
 
 | |
7OA6
 
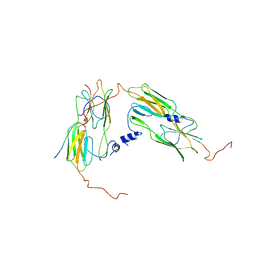 | | Pseudo-atomic model for Hsp26 residues 63 to 214. Please be advised that the target map is not of sufficient resolution to unambiguously position backbone or side chain atoms. This model represents a likely fit. | | Descriptor: | Heat shock protein 26 | | Authors: | Muehlhofer, M, Peters, C, Kriehuber, T, Kreuzeder, M, Kazman, P, Rodina, N, Reif, B, Haslbeck, M, Weinkauf, S, Buchner, J. | | Deposit date: | 2021-04-19 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Phosphorylation activates the yeast small heat shock protein Hsp26 by weakening domain contacts in the oligomer ensemble.
Nat Commun, 12, 2021
|
|
8GJY
 
 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | Descriptor: | cGAS-like receptor 1 | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
4L53
 
 | | Crystal Structure of (1R,4R)-4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)-3-chlorofuro[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}cyclohexan-1-ol bound to TAK1-TAB1 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, ... | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Jestel, A, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1: Optimization of kinase selectivity and pharmacokinetics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7O6P
 
 | |
3ZSN
 
 | | Structure of the mixed-function P450 MycG F286A mutant in complex with mycinamicin IV | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Rutaganira, F.U, Anzai, Y, Kato, F, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-06-29 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
1XL0
 
 | | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1,3,4-oxadiazole,-benzothiazole, and-benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site. | | Descriptor: | (1R)-1,5-anhydro-1-(5-methyl-1,3,4-oxadiazol-2-yl)-D-glucitol, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Tiraidis, C, Kardakaris, R, Bischler, N, Leonidas, D.D, Hadady, Z, Somsak, L, Docsa, T, Gergely, P, Oikonomakos, N.G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1, 3, 4-oxadiazole, -benzothiazole, and -benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site
Protein Sci., 14, 2005
|
|
