2VVY
 
 | | Structure of Vaccinia virus protein B14 | | Descriptor: | PROTEIN B15 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2V8O
 
 | | Structure of the Murray Valley encephalitis virus RNA helicase to 1. 9A resolution | | Descriptor: | FLAVIVIRIN PROTEASE NS3 | | Authors: | Mancini, E.J, Assenberg, R, Verma, A, Walter, T.S, Tuma, R, Grimes, J.M, Owens, R.J, Stuart, D.I. | | Deposit date: | 2007-08-09 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Murray Valley Encephalitis Virus RNA Helicase at 1.9 A Resolution.
Protein Sci., 16, 2007
|
|
2UVD
 
 | | The crystal structure of a 3-oxoacyl-(acyl carrier protein) reductase from Bacillus anthracis (BA3989) | | Descriptor: | 3-OXOACYL-(ACYL-CARRIER-PROTEIN) REDUCTASE | | Authors: | Zaccai, N.R, Carter, L.G, Berrow, N.S, Sainsbury, S, Nettleship, J.E, Walter, T.S, Harlos, K, Owens, R.J, Wilson, K.S, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a 3-Oxoacyl-(Acylcarrier Protein) Reductase (Ba3989) from Bacillus Anthracis at 2.4-A Resolution.
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
2VVX
 
 | | Structure of Vaccinia virus protein A52 | | Descriptor: | PROTEIN A52 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2VD8
 
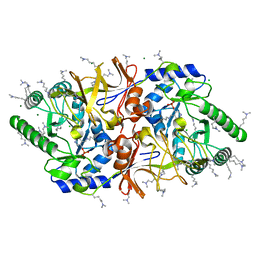 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) | | Descriptor: | ALANINE RACEMASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2W2R
 
 | | Structure of the vesicular stomatitis virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
2WO2
 
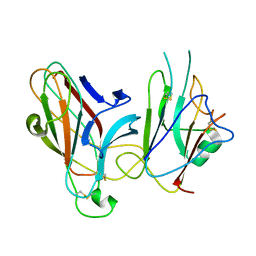 | | Crystal Structure of the EphA4-ephrinB2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EPHRIN TYPE-A RECEPTOR, EPHRIN-B2 | | Authors: | Bowden, T.A, Aricescu, A.R, Nettleship, J.E, Siebold, C, Rahman-Huq, N, Owens, R.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2009-07-21 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Plasticity of Eph-Receptor A4 Facilitates Cross-Class Ephrin Signalling
Structure, 17, 2009
|
|
2WO3
 
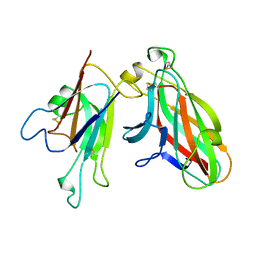 | | Crystal Structure of the EphA4-ephrinA2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPHRIN TYPE-A RECEPTOR, EPHRIN-A2 | | Authors: | Bowden, T.A, Aricescu, A.R, Nettleship, J.E, Siebold, C, Rahman-Huq, N, Owens, R.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2009-07-21 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Plasticity of Eph-Receptor A4 Facilitates Cross-Class Ephrin Signalling
Structure, 17, 2009
|
|
2GW8
 
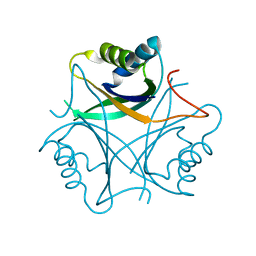 | | Structure of the PII signal transduction protein of Neisseria meningitidis at 1.85 resolution | | Descriptor: | PII signal transduction protein | | Authors: | Nichols, C.E, Sainsbury, S, Berrow, N.S, Alderton, D, Stammers, D.K, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the P(II) signal transduction protein of Neisseria meningitidis at 1.85 A resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2JCB
 
 | | The crystal structure of 5-formyl-tetrahydrofolate cycloligase from Bacillus anthracis (BA4489) | | Descriptor: | 5-FORMYLTETRAHYDROFOLATE CYCLO-LIGASE FAMILY PROTEIN, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Meier, C, Carter, L.G, Winter, G, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-21 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of 5-Formyltetrahydrofolate Cyclo-Ligase from Bacillus Anthracis (Ba4489).
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2JJX
 
 | | THE CRYSTAL STRUCTURE OF UMP KINASE FROM BACILLUS ANTHRACIS (BA1797) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, URIDYLATE KINASE | | Authors: | Meier, C, Carter, L.G, Mancini, E.J, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Crystal Structure of Ump Kinase from Bacillus Anthracis (Ba1797) Reveals an Allosteric Nucleotide-Binding Site.
J.Mol.Biol., 381, 2008
|
|
8OWV
 
 | | H6 and F2 nanobodies bound to SARS-CoV-2 spike RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2, GLYCEROL, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OYU
 
 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OYT
 
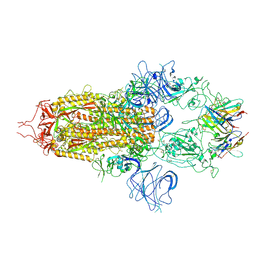 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWT
 
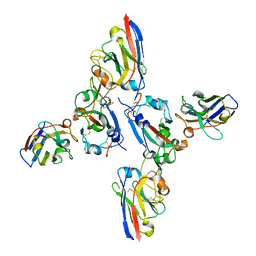 | | SARS-CoV-2 spike RBD with A8 and H3 nanobodies bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A8, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWW
 
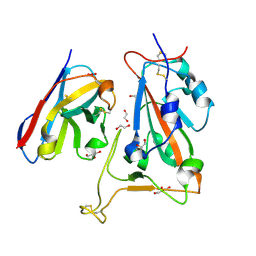 | | B5-5 nanobody bound to SARS-CoV-2 spike RBD (Wuhan) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, B5-5 nanobody, ... | | Authors: | Cornish, K.A.S, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
3X1O
 
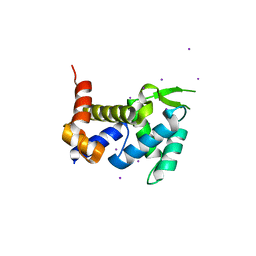 | | Crystal structure of the ROQ domain of human Roquin | | Descriptor: | IODIDE ION, Roquin-1 | | Authors: | Ose, T, Verma, A, Cockburn, J.B, Berrow, N.S, Alderton, D, Stuart, D, Owens, R.J, Jones, E.Y. | | Deposit date: | 2014-11-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Roquin binds microRNA-146a and Argonaute2 to regulate microRNA homeostasis
Nat Commun, 6, 2015
|
|
8PX4
 
 | | Structure of the PAS domain code by the LIC_11128 gene from Leptospira interrogans serovar Copenhageni Fiocruz, solved at wavelength 3.09 A | | Descriptor: | Diguanylate cyclase | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Guzzo, C.R, Owens, R.J, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8P6H
 
 | | Bovine naive ultralong antibody AbBLV5B8* collected at 100K | | Descriptor: | Antibody BLV5B8* heavy chain, Antibody BLV5B8* light chain | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2023-05-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The impact of chain-exchange on bovine ultralong immunoglobulins.
Acta Crystallographica Section F, 2023
|
|
8P2T
 
 | | Bovine naive ultralong antibody AbD08* collected at 100K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Antibody D08* heavy chain, Antibody D08* light chain, ... | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The impact of chain-exchange on bovine ultralong immunoglobulins.
Acta Crystallographica Section F, 2024
|
|
8PDD
 
 | | Thioredoxin glutathione reductase of Schistosoma mansoni at 1.25A resolution. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ribeiro, L, Montoya, B.O, Moreira-Filho, J.T, Bowyer, S, Verma, A, Neves, B.J, Owens, R.J, Andrade, C.H, Silva-Jr, F.P, Furnham, N. | | Deposit date: | 2023-06-12 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni.
Sci Rep, 14, 2024
|
|
8PL7
 
 | | Thioredoxin glutathione reductase of Schistosoma mansoni fragment screen hit 8. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase, ~{N}-morpholin-4-ylcarbothioylfuran-2-carboxamide | | Authors: | Ribeiro, L, Montoya, B.O, Moreira-Filho, J.T, Bowyer, S, Verma, A, Neves, B.J, Owens, R.J, Andrade, C.H, Silva-Jr, F.P, Furnham, N. | | Deposit date: | 2023-06-27 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni.
Sci Rep, 14, 2024
|
|
8PLQ
 
 | | Thioredoxin glutathione reductase of Schistosoma mansoni fragment screen hit 27. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase, ~{N}-(1-ethylbenzimidazol-2-yl)ethanamide | | Authors: | Ribeiro, L, Montoya, B.O, Moreira-Filho, J.T, Bowyer, S, Verma, A, Neves, B.J, Owens, R.J, Andrade, C.H, Silva-Jr, F.P, Furnham, N. | | Deposit date: | 2023-06-27 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni.
Sci Rep, 14, 2024
|
|
8PLM
 
 | | Thioredoxin glutathione reductase of Schistosoma mansoni fragment screen hit 23. | | Descriptor: | 1-{4-[(2-methoxyethyl)amino]piperidin-1-yl}ethan-1-one, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | Authors: | Ribeiro, L, Montoya, B.O, Moreira-Filho, J.T, Bowyer, S, Verma, A, Neves, B.J, Owens, R.J, Andrade, C.H, Silva-Jr, F.P, Furnham, N. | | Deposit date: | 2023-06-27 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni.
Sci Rep, 14, 2024
|
|
8PLX
 
 | | Thioredoxin glutathione reductase of Schistosoma mansoni fragment screen hit 34. | | Descriptor: | 4-(4-fluorophenyl)piperazine-1-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | Authors: | Ribeiro, L, Montoya, B.O, Moreira-Filho, J.T, Bowyer, S, Verma, A, Neves, B.J, Owens, R.J, Andrade, C.H, Silva-Jr, F.P, Furnham, N. | | Deposit date: | 2023-06-27 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni.
Sci Rep, 14, 2024
|
|
