7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
3OC2
 
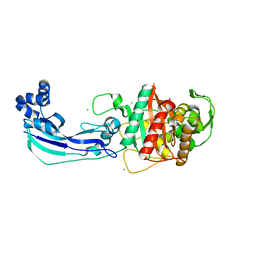 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Penicillin-binding protein 3 | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
2WV9
 
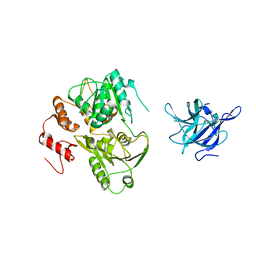 | | Crystal Structure of the NS3 protease-helicase from Murray Valley encephalitis virus | | Descriptor: | FLAVIVIRIN PROTEASE NS2B REGULATORY SUBUNIT, FLAVIVIRIN PROTEASE NS3 CATALYTIC SUBUNIT | | Authors: | Assenberg, R, Mastrangelo, E, Walter, T.S, Verma, A, Milani, M, Owens, R.J, Stuart, D.I, Grimes, J.M, Mancini, E.J. | | Deposit date: | 2009-10-15 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of a Novel Conformational State of the Flavivirus Ns3 Protein: Implications for Polyprotein Processing and Viral Replication.
J.Virol., 83, 2009
|
|
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZAW
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
2WO1
 
 | | Crystal Structure of the EphA4 Ligand Binding Domain | | Descriptor: | EPHRIN TYPE-A RECEPTOR, N-PROPANOL | | Authors: | Bowden, T.A, Aricescu, A.R, Nettleship, J.E, Siebold, C, Rahman-Huq, N, Owens, R.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2009-07-21 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Plasticity of Eph-Receptor A4 Facilitates Cross-Class Ephrin Signalling
Structure, 17, 2009
|
|
6QNA
 
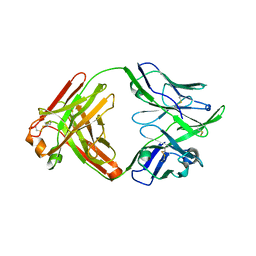 | | Structure of bovine anti-RSV hybrid Fab B13HC-B4LC | | Descriptor: | B13 Heavy chain, B4 light chain, GLYCEROL | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN9
 
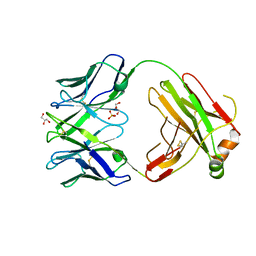 | | Structure of bovine anti-RSV Fab B4 | | Descriptor: | GLYCEROL, Heavy chain, SULFATE ION, ... | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN7
 
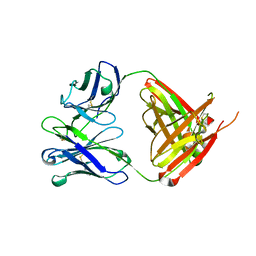 | | Structure of bovine anti-RSV hybrid Fab B4HC-B13LC | | Descriptor: | Heavy chain of bovine anti-RSV B4, Light chain of bovine anti-RSV B13 | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
4AV2
 
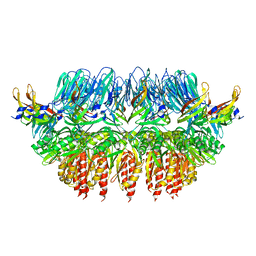 | | Single particle electron microscopy of PilQ dodecameric complexes from Neisseria meningitidis. | | Descriptor: | PILP PROTEIN, TYPE IV PILUS BIOGENESIS AND COMPETENCE PROTEIN PILQ | | Authors: | Berry, J.L, Phelan, M.M, Collins, R.F, Adomavicius, T, Tonjum, T, Frye, S.A, Bird, L, Owens, R, Ford, R.C, Lian, L.Y, Derrick, J.P. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Structure and Assembly of a Trans-Periplasmic Channel for Type Iv Pili in Neisseria Meningitidis.
Plos Pathog., 8, 2012
|
|
4BBK
 
 | | Structural and functional characterisation of the kindlin-1 pleckstrin homology domain | | Descriptor: | FERMITIN FAMILY HOMOLOG 1, GLYCEROL | | Authors: | Yates, L.A, Lumb, C.N, Brahme, N.N, Zalyte, R, Bird, L.E, De Colibus, L, Owens, R.J, Calderwood, D.A, Sansom, M.S.P, Gilbert, R.J.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterisation of the Kindlin-1 Pleckstrin Homology Domain
J.Biol.Chem., 287, 2012
|
|
4YU8
 
 | | Crystal structure of Neuroblastoma suppressor of tumorigenicity 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Neuroblastoma suppressor of tumorigenicity 1 | | Authors: | Ren, J, Nettleship, J.E, Stammers, D.K, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2015-03-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Neuroblastoma suppressor of tumorigenicity 1
To Be Published
|
|
5ANQ
 
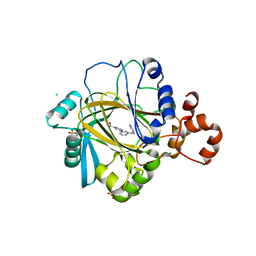 | | inhibitors of JumonjiC domain-containing histone demethylases | | Descriptor: | 2-{2-[(pyridin-3-ylmethyl)amino]pyrimidin-4-yl}pyridine-4-carboxylic acid, CHLORIDE ION, FE (II) ION, ... | | Authors: | Roatsch, M, Robaa, D, Pippel, M, Nettleship, J.E, Reddivari, Y, Bird, L.E, Hoffmann, I, Franz, H, Owens, R.J, Schuele, R, Flaig, R, Sippl, W, Jung, M. | | Deposit date: | 2015-09-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substituted 2-(2-Aminopyrimidin-4-Yl)Pyridine-4-Carboxylates as Potent Inhibitors of Jumonjic Domain-Containing Histone Demethylases.
Fut.Med.Chem., 8, 2016
|
|
3CAM
 
 | |
3DGG
 
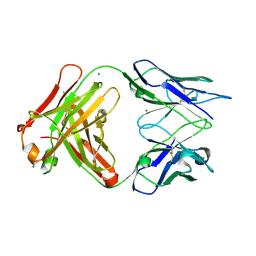 | | Crystal structure of FabOX108 | | Descriptor: | FabOX108 Heavy Chain Fragment, FabOX108 Light Chain Fragment, MAGNESIUM ION | | Authors: | Ren, J, Nettleship, J.E, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2008-06-13 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A pipeline for the production of antibody fragments for structural studies using transient expression in HEK 293T cells.
Protein Expr.Purif., 62, 2008
|
|
5EYT
 
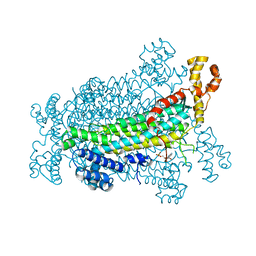 | | Crystal Structure of Adenylosuccinate Lyase from Schistosoma mansoni in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase | | Authors: | Romanello, L, Torini, J.R, Bird, L, Nettleship, J, Owens, R, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3649 Å) | | Cite: | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
3G3Z
 
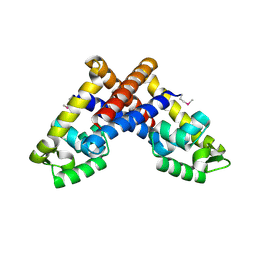 | | The structure of NMB1585, a MarR family regulator from Neisseria meningitidis | | Descriptor: | Transcriptional regulator, MarR family | | Authors: | Nichols, C.E, Sainsbury, S, Ren, J, Walter, T.S, Verma, A, Stammers, D.K, Saunders, N.J, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of NMB1585, a MarR-family regulator from Neisseria meningitidis
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3DIF
 
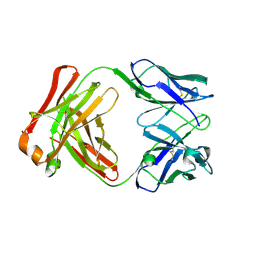 | |
4AB6
 
 | | Regulatory domain structure of NMB2055 (MetR), C103S C106S mutant, a LysR family regulator from N. meningitidis | | Descriptor: | SULFATE ION, TRANSCRIPTIONAL REGULATOR, LYSR FAMILY | | Authors: | Sainsbury, S, Ren, J, Saunders, N.J, Stuart, D.I, Owens, R.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Regulatory Domain of the Lysr Family Regulator Nmb2055 (Metr-Like Protein) from Neisseria Meningitidis
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4AB5
 
 | | Regulatory domain structure of NMB2055 (MetR) a LysR family regulator from N. meningitidis | | Descriptor: | TRANSCRIPTIONAL REGULATOR, LYSR FAMILY | | Authors: | Sainsbury, S, Ren, J, Saunders, N.J, Stuart, D.I, Owens, R.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the Regulatory Domain of the Lysr Family Regulator Nmb2055 (Metr-Like Protein) from Neisseria Meningitidis
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3HHF
 
 | | Structure of CrgA regulatory domain, a LysR-type transcriptional regulator from Neisseria meningitidis. | | Descriptor: | CHLORIDE ION, Transcriptional regulator, LysR family | | Authors: | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
3HHG
 
 | | Structure of CrgA, a LysR-type transcriptional regulator from Neisseria meningitidis. | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
5A9D
 
 | | Crystal structure of the extracellular domain of PepT1 | | Descriptor: | GLYCEROL, SOLUTE CARRIER FAMILY 15 MEMBER 1 | | Authors: | Beale, J.H, Bird, L.E, Owens, R.J, Newstead, S. | | Deposit date: | 2015-07-20 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Extracellular Domain from Pept1 and Pept2 Provide Novel Insights Into Mammalian Peptide Transport
Structure, 23, 2015
|
|
