4PKD
 
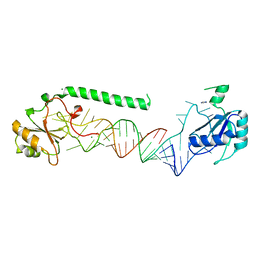 | | U1-70k in complex with U1 snRNA stem-loops 1 and U1-A RRM in complex with stem-loop 2 | | Descriptor: | IMIDAZOLE, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A,U1 small nuclear ribonucleoprotein 70 kDa, ... | | Authors: | Oubridge, C, Kondo, Y, van Roon, A.M, Nagai, K. | | Deposit date: | 2014-05-14 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5' splice site recognition.
Elife, 4, 2015
|
|
1URN
 
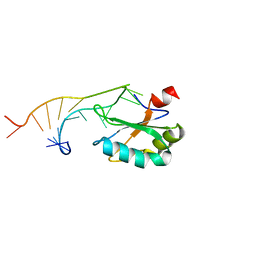 | | U1A MUTANT/RNA COMPLEX + GLYCEROL | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTEIN (U1A), ... | | Authors: | Oubridge, C, Ito, N, Evans, P.R, Teo, C.-H, Nagai, K. | | Deposit date: | 1995-01-04 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin.
Nature, 372, 1994
|
|
1L9A
 
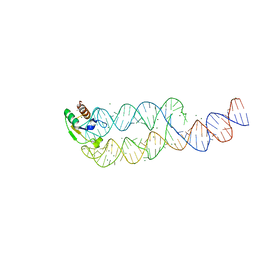 | | CRYSTAL STRUCTURE OF SRP19 IN COMPLEX WITH THE S DOMAIN OF SIGNAL RECOGNITION PARTICLE RNA | | Descriptor: | MAGNESIUM ION, METHYL MERCURY ION, SIGNAL RECOGNITION PARTICLE 19 KDA PROTEIN, ... | | Authors: | Oubridge, C, Kuglstatter, A, Jovine, L, Nagai, K. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle.
Mol.Cell, 9, 2002
|
|
3ZEF
 
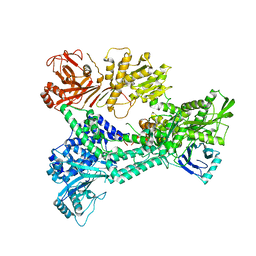 | | Crystal structure of Prp8:Aar2 complex: second crystal form at 3.1 Angstrom resolution | | Descriptor: | A1 CISTRON-SPLICING FACTOR AAR2, PRE-MRNA-SPLICING FACTOR 8 | | Authors: | Galej, W.P, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Prp8 Reveals Active Site Cavity of the Spliceosome
Nature, 493, 2013
|
|
3KL4
 
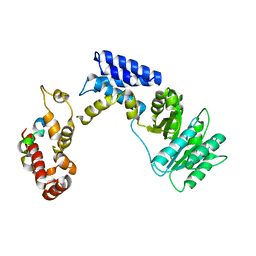 | | Recognition of a signal peptide by the signal recognition particle | | Descriptor: | Signal peptide of yeast dipeptidyl aminopeptidase B, Signal recognition 54 kDa protein | | Authors: | Janda, C.Y, Nagai, K, Li, J, Oubridge, C. | | Deposit date: | 2009-11-06 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Recognition of a signal peptide by the signal recognition particle.
Nature, 465, 2010
|
|
3CW1
 
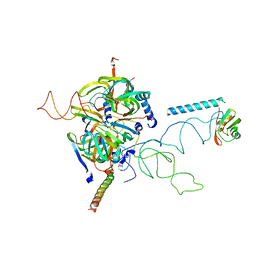 | | Crystal Structure of Human Spliceosomal U1 snRNP | | Descriptor: | Small nuclear ribonucleoprotein E, Small nuclear ribonucleoprotein F, Small nuclear ribonucleoprotein G, ... | | Authors: | Pomeranz Krummel, D.A, Oubridge, C, Leung, A.K, Li, J, Nagai, K. | | Deposit date: | 2008-04-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.493 Å) | | Cite: | Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution.
Nature, 458, 2009
|
|
6QDV
 
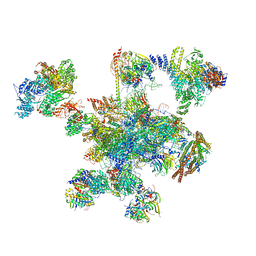 | | Human post-catalytic P complex spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Fica, S.M, Oubridge, C, Wilkinson, M.E, Newman, A.J, Nagai, K. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation.
Science, 363, 2019
|
|
4I43
 
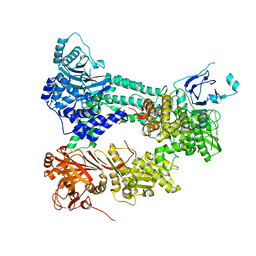 | | Crystal structure of Prp8:Aar2 complex | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Galej, W.P, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2012-11-27 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Prp8 reveals active site cavity of the spliceosome.
Nature, 493, 2013
|
|
5LSB
 
 | | Crystal structure of yeast Hsh49p in complex with Cus1p binding domain. | | Descriptor: | Cold sensitive U2 snRNA suppressor 1, Protein HSH49 | | Authors: | van Roon, A.M, Obayashi, E, Sposito, B, Oubridge, C, Nagai, K. | | Deposit date: | 2016-08-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain.
RNA, 23, 2017
|
|
1DUH
 
 | | CRYSTAL STRUCTURE OF THE CONSERVED DOMAIN IV OF E. COLI 4.5S RNA | | Descriptor: | 4.5S RNA DOMAIN IV, LUTETIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Jovine, L, Hainzl, T, Oubridge, C, Scott, W.G, Li, J, Sixma, T.K, Wonacott, A, Skarzynski, T, Nagai, K. | | Deposit date: | 2000-01-17 | | Release date: | 2000-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA.
Structure Fold.Des., 8, 2000
|
|
5LJ3
 
 | | Structure of the core of the yeast spliceosome immediately after branching | | Descriptor: | CEF1, CLF1, CWC15, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
2VRD
 
 | | THE STRUCTURE OF THE ZINC FINGER FROM THE HUMAN SPLICEOSOMAL PROTEIN U1C | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN C, ZINC ION | | Authors: | Muto, Y, Pomeranz-Krummel, D, Oubridge, C, Hernandez, H, Robinson, C, Neuhaus, D, Nagai, K. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure and Biochemical Properties of the Human Spliceosomal Protein U1C
J.Mol.Biol., 341, 2004
|
|
5MPS
 
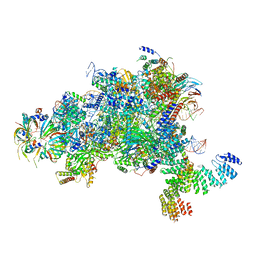 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-18 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
4PJO
 
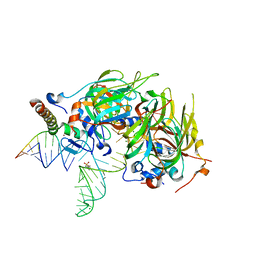 | | Minimal U1 snRNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ETHANOL, ... | | Authors: | Kondo, Y, Oubridge, C, van Roon, A.M, Nagai, K. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5' splice site recognition.
Elife, 4, 2015
|
|
1MFQ
 
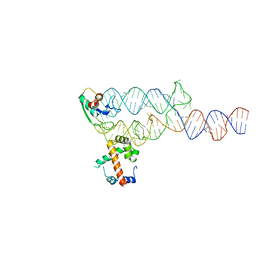 | | Crystal Structure Analysis of a Ternary S-Domain Complex of Human Signal Recognition Particle | | Descriptor: | 7S RNA of human SRP, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kuglstatter, A, Oubridge, C, Nagai, K. | | Deposit date: | 2002-08-13 | | Release date: | 2002-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Induced structural changes of 7SL RNA during the assembly of human
signal recognition particle
Nat.Struct.Biol., 9, 2002
|
|
5LSL
 
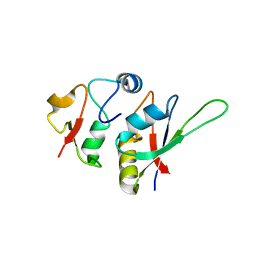 | | Crystal structure of yeast Hsh49p in complex with Cus1p binding domain. | | Descriptor: | Cold sensitive U2 snRNA suppressor 1, Protein HSH49 | | Authors: | van Roon, A.M, Obayashi, E, Sposito, B, Oubridge, C, Nagai, K. | | Deposit date: | 2016-09-02 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of U2 snRNP SF3b components: Hsh49p in complex with Cus1p-binding domain.
RNA, 23, 2017
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | Descriptor: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
5GAM
 
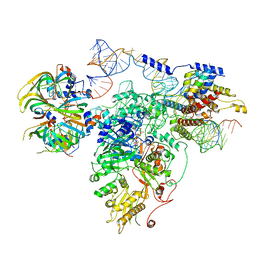 | | Foot region of the yeast spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Pre-mRNA-splicing factor 8, Pre-mRNA-splicing factor SNU114, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Angstrom resolution
Nature, 530, 2016
|
|
5GAO
 
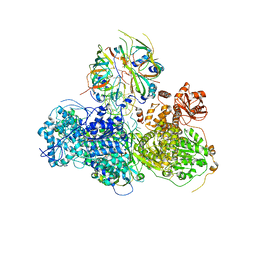 | | Head region of the yeast spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | Pre-mRNA-splicing factor 8, Pre-mRNA-splicing helicase BRR2, Saccharomyces cerevisiae strain UOA_M2 chromosome 5 sequence, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
5GAP
 
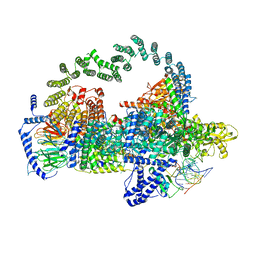 | | Body region of the U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, Pre-mRNA-processing factor 31, Pre-mRNA-splicing factor 6, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Oubridge, C, Bai, X.C, Newman, A, Scheres, S, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
5GAN
 
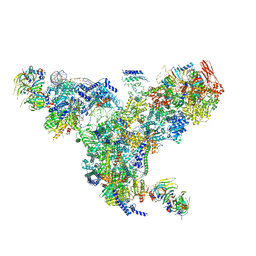 | | The overall structure of the yeast spliceosomal U4/U6.U5 tri-snRNP at 3.7 Angstrom | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, Pre-mRNA-processing factor 31, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
