6YMX
 
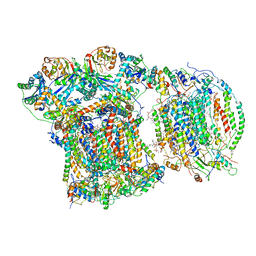 | | CIII2/CIV respiratory supercomplex from Saccharomyces cerevisiae | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (1R)-2-(phosphonooxy)-1-[(tridecanoyloxy)methyl]ethyl pentadecanoate, (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, ... | | Authors: | Berndtsson, J, Rathore, S, Ott, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Embo Rep., 21, 2020
|
|
6YMY
 
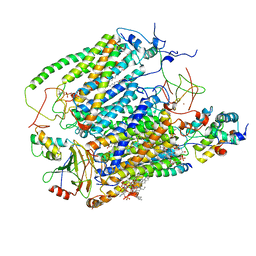 | | Cytochrome c oxidase from Saccharomyces cerevisiae | | Descriptor: | (2R,5S,11R,14R)-5,8,11-trihydroxy-2-(nonanoyloxy)-5,11-dioxido-16-oxo-14-[(propanoyloxy)methyl]-4,6,10,12,15-pentaoxa-5,11-diphosphanonadec-1-yl undecanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, COPPER (II) ION, ... | | Authors: | Berndtsson, J, Rathore, S, Ott, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Embo Rep., 21, 2020
|
|
1JM4
 
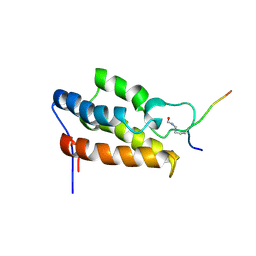 | | NMR Structure of P/CAF Bromodomain in Complex with HIV-1 Tat Peptide | | Descriptor: | HIV-1 Tat Peptide, P300/CBP-associated Factor | | Authors: | Mujtaba, S, He, Y, Zeng, L, Farooq, A, Carlson, J.E, Ott, M, Verdin, E, Zhou, M.-M. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain
Mol.Cell, 9, 2002
|
|
2MZB
 
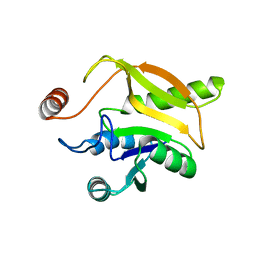 | | Solution structural studies of GTP:adenosylcobinamide-phosphate guanylyltransferase (CobY) from Methanocaldococcus jannaschii | | Descriptor: | Adenosylcobinamide-phosphate guanylyltransferase | | Authors: | Singarapu, K, Otte, M, Tonelli, M, Westler, W.M, Escalante-Semerena, J.C, Markley, J.L. | | Deposit date: | 2015-02-11 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structural Studies of GTP:Adenosylcobinamide-Phosphateguanylyl Transferase (CobY) from Methanocaldococcus jannaschii.
Plos One, 10, 2015
|
|
6URE
 
 | | Barrier-to-autointegration factor Aqueous: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URK
 
 | | Barrier-to-autointegration factor soaked in Glycerol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6USD
 
 | | Barrier-to-autointegration factor soaked in ethanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URJ
 
 | | Barrier-to-autointegration factor soaked in Acetone: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6USB
 
 | | Barrier-to-autointegration factor soaked in urea: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US1
 
 | | Barrier-to-autointegration factor soaked in isobutanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US7
 
 | | Barrier-to-autointegration factor soaked in TMAO: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URL
 
 | | Barrier-to-autointegration factor soaked in isopropanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URR
 
 | | Barrier-to-autointegration factor soaked in Dioxane: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6USI
 
 | | Barrier-to-autointegration factor soaked in 1,6-hexanediol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URZ
 
 | | Barrier-to-autointegration factor soaked in methanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL, METHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URN
 
 | | Barrier-to-autointegration factor t-butanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US0
 
 | | Barrier-to-autointegration factor soaked in R,S,R-bisfuranol (RSR): 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6UNT
 
 | | Barrier-to-autointegration factor soaked in DMSO: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-13 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4TZG
 
 | | Crystal structure of eCGP123, an extremely thermostable green fluorescent protein | | Descriptor: | Fluorescent Protein | | Authors: | Close, D.W, Don Paul, C, Traore, D.A.K, Wilce, M.C.J, Prescott, M, Bradbury, A.R.M. | | Deposit date: | 2014-07-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermal green protein, an extremely stable, nonaggregating fluorescent protein created by structure-guided surface engineering.
Proteins, 83, 2015
|
|
6CBE
 
 | | Atomic structure of a rationally engineered gene delivery vector, AAV2.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Burg, M, Rosebrough, C, Drouin, L, Bennett, A, Mietzsch, M, Chipman, P, McKenna, R, Sousa, D, Potter, M, Byrne, B, Kozyreva, O.G, Samulski, R.J, Agbandje-McKenna, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Atomic structure of a rationally engineered gene delivery vector, AAV2.5.
J. Struct. Biol., 203, 2018
|
|
7S8Z
 
 | |
7S9G
 
 | | Room-temperature Human Hsp90a-NTD bound to BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Stachowski, T.R, Vanarotti, M, Fischer, M. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7S9F
 
 | | Cryogenic Human Hsp90a-NTD bound to BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Stachowski, T.R, Vanarotti, M, Seetharaman, J, Fischer, M. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7S95
 
 | | Room-temperature Human Hsp90a-NTD bound to adenine | | Descriptor: | ADENINE, Heat shock protein HSP 90-alpha | | Authors: | Stachowski, T.R, Vanarotti, M, Lopez, K, Fischer, M. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7S9I
 
 | | Room-temperature Human Hsp90a-NTD bound to EC144 | | Descriptor: | 5-{2-amino-4-chloro-7-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-7H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-methylpent-4-yn-2 -ol, Heat shock protein HSP 90-alpha | | Authors: | Stachowski, T.R, Vanarotti, M, Fischer, M. | | Deposit date: | 2021-09-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Water Networks Repopulate Protein-Ligand Interfaces with Temperature.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
