7F1P
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant ligand-free form. | | Descriptor: | L-methionine gamma-lyase | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7X7X
 
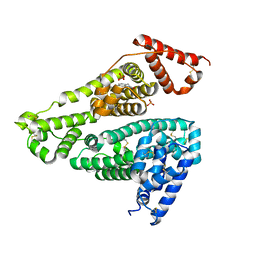 | | Human serum albumin complex with deschloro-aripiprazole | | Descriptor: | 7-[4-(4-phenylpiperazin-1-yl)butoxy]-3,4-dihydro-1H-quinolin-2-one, PHOSPHATE ION, Serum albumin | | Authors: | Kawai, A, Otagiri, M. | | Deposit date: | 2022-03-10 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chlorine Atoms of an Aripiprazole Molecule Control the Geometry and Motion of Aripiprazole and Deschloro-aripiprazole in Subdomain IIIA of Human Serum Albumin.
Acs Omega, 7, 2022
|
|
7E3N
 
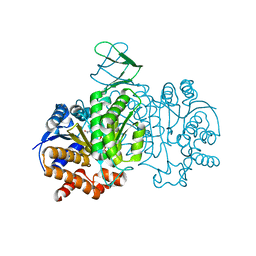 | | Crystal structure of Isocitrate dehydrogenase D252N mutant from Trypanosoma brucei in complexed with NADP+, alpha-ketoglutarate, and ca2+ | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], ... | | Authors: | Arai, N, Shiba, T, Inaoka, D.K, Kita, K, Wang, X, Otani, M, Matsushiro, S, Kojima, C. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Isocitrate dehydrogenase from Trypanosoma brucei.
To Be Published
|
|
3MMR
 
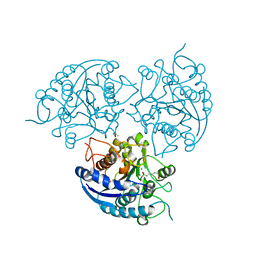 | | Structure of Plasmodium falciparum Arginase in complex with ABH | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase, BETA-MERCAPTOETHANOL, ... | | Authors: | Dowling, D.P, Ilies, M, Olszewski, K.L, Portugal, S, Mota, M.M, Llinas, M, Christianson, D.W. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of arginase from Plasmodium falciparum and implications for L-arginine depletion in malarial infection .
Biochemistry, 49, 2010
|
|
6A7P
 
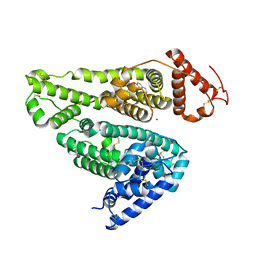 | | Human serum albumin complexed with aripiprazole | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kawai, A, Yamasaki, K, Otagiri, M. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Analysis of the Binding of Aripiprazole to Human Serum Albumin: The Importance of a Chloro-Group in the Chemical Structure.
Acs Omega, 3, 2018
|
|
5BNF
 
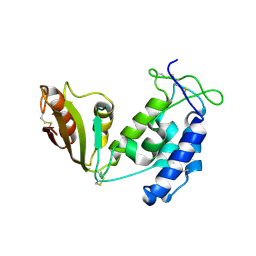 | | Apo structure of porcine CD38 | | Descriptor: | Uncharacterized protein | | Authors: | Ting, K.Y, Leung, C.P.F, Graeff, R.M, Lee, H.C, Hao, Q, Kotaka, M. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Porcine CD38 exhibits prominent secondary NAD(+) cyclase activity.
Protein Sci., 25, 2016
|
|
5BNI
 
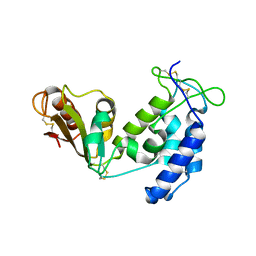 | | Porcine CD38 complexed with complexed with a covalent intermediate, ribo-F-ribose-5'-phosphate | | Descriptor: | Uncharacterized protein, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4S)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Ting, K.Y, Leung, C.F.P, Graeff, R.M, Lee, H.C, Hao, Q, Kotaka, M. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Porcine CD38 exhibits prominent secondary NAD(+) cyclase activity.
Protein Sci., 25, 2016
|
|
5X52
 
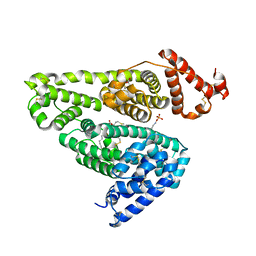 | | Human serum albumin complexed with octanoate and N-acetyl-L-methionine | | Descriptor: | N-ACETYLMETHIONINE, OCTANOIC ACID (CAPRYLIC ACID), PHOSPHATE ION, ... | | Authors: | Kawai, A, Otagiri, M. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Crystallographic analysis of the ternary complex of octanoate and N-acetyl-l-methionine with human serum albumin reveals the mode of their stabilizing interactions
Biochim. Biophys. Acta, 1865, 2017
|
|
6A2U
 
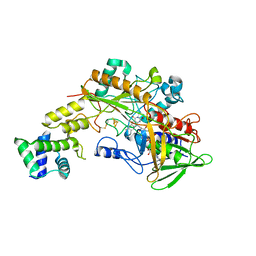 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Glucose dehydrogenase, ... | | Authors: | Yoshida, H, Kojima, K, Yoshimatsu, K, Shiota, M, Yamazaki, T, Ferri, S, Tsugawa, W, Kamitori, S, Sode, K. | | Deposit date: | 2018-06-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the direct electron transfer-type FAD glucose dehydrogenase catalytic subunit complexed with a hitchhiker protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1B5X
 
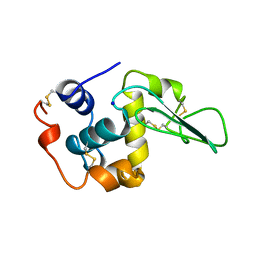 | | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and x-ray analysis of six ser->ala mutants | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5U
 
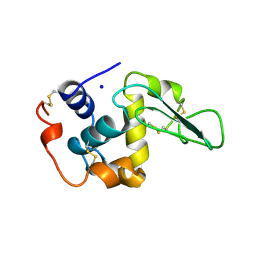 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANT | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5Z
 
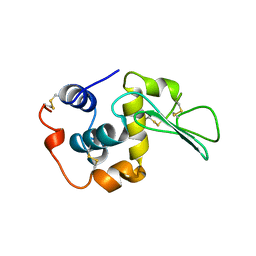 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | LYSOZYME | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5W
 
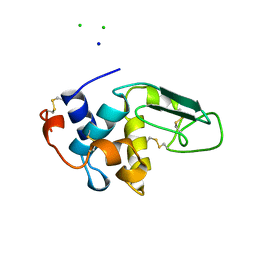 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5V
 
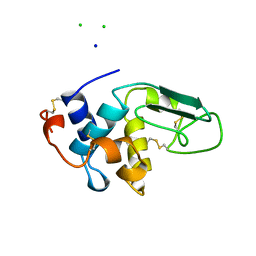 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
3SY8
 
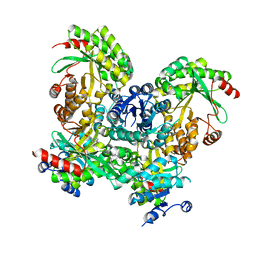 | | Crystal structure of the response regulator RocR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RocR | | Authors: | Chen, M.W, Kotaka, M, Vonrhein, C, Bricogne, G, Lescar, J. | | Deposit date: | 2011-07-16 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the regulatory mechanism of the response regulator RocR from Pseudomonas aeruginosa in cyclic Di-GMP signaling.
J.Bacteriol., 194, 2012
|
|
1B5Y
 
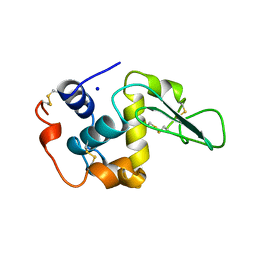 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1D7Y
 
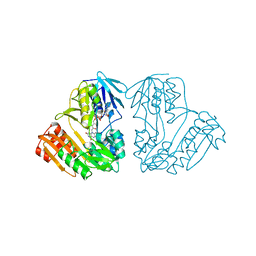 | | CRYSTAL STRUCTURE OF NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4 | | Descriptor: | FERREDOXIN REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Senda, T, Yamada, T, Sakurai, N, Kubota, M, Nishizaki, T, Masai, E, Fukuda, M, Mitsui, Y. | | Deposit date: | 1999-10-21 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase.
J.Mol.Biol., 304, 2000
|
|
5YOQ
 
 | |
5YXW
 
 | | Crystal structure of the prefusion form of measles virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, glycoprotein F1,measles virus fusion protein, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-07 | | Release date: | 2018-02-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.776 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YZD
 
 | | Crystal structure of the prefusion form of measles virus fusion protein in complex with a fusion inhibitor peptide (FIP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, glycoprotein F1,measles virus fusion protein, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.636 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YZC
 
 | | Crystal structure of the prefusion form of measles virus fusion protein in complex with a fusion inhibitor compound (AS-48) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitro-2-[(phenylacetyl)amino]benzamide, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1BF2
 
 | | STRUCTURE OF PSEUDOMONAS ISOAMYLASE | | Descriptor: | CALCIUM ION, ISOAMYLASE | | Authors: | Katsuya, Y, Mezaki, Y, Kubota, M, Matsuura, Y. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Pseudomonas isoamylase at 2.2 A resolution.
J.Mol.Biol., 281, 1998
|
|
3I9O
 
 | | Crystal structure of ADP ribosyl cyclase complexed with ribo-2'F-ADP ribose | | Descriptor: | ADP-ribosyl cyclase, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4S)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Graeff, R, Liu, Q, Kriksunov, I.A, Kotaka, M, Oppenheimer, N, Hao, Q, Lee, H.C. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of cyclizing NAD to cyclic ADP-ribose by ADP-ribosyl cyclase and CD38
J.Biol.Chem., 284, 2009
|
|
3ZH2
 
 | | Structure of Plasmodium falciparum lactate dehydrogenase in complex with a DNA aptamer | | Descriptor: | DNA APTAMER, L-LACTATE DEHYDROGENASE | | Authors: | Cheung, Y.W, Kwok, J, Law, A.W.L, Watt, R.M, Kotaka, M, Tanner, J.A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Discriminatory Recognition of Plasmodium Lactate Dehydrogenase by a DNA Aptamer
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3K1I
 
 | | Crystal strcture of FliS-HP1076 complex in H. pylori | | Descriptor: | Flagellar protein, Putative uncharacterized protein | | Authors: | Lam, W.W.L, Kotaka, M, Ling, T.K.W, Au, S.W.N. | | Deposit date: | 2009-09-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular interaction of flagellar export chaperone FliS and cochaperone HP1076 in Helicobacter pylori
Faseb J., 24, 2010
|
|
