8XGR
 
 | | ETB-eGt complex bound to endothelin-1 | | Descriptor: | Camelid antibody VHH fragment, Endothelin receptor type B, Endothelin-1, ... | | Authors: | Oshima, H.S, Sano, F.K, Akasaka, H, Iwama, A, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-15 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Optimizing cryo-EM structural analysis of G i -coupling receptors via engineered G t and Nb35 application.
Biochem.Biophys.Res.Commun., 693, 2024
|
|
1GC0
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
1GC2
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
1RFP
 
 | |
1KXW
 
 | |
1KXX
 
 | |
1KXY
 
 | |
1J1T
 
 | | Alginate lyase from Alteromonas sp.272 | | Descriptor: | Alginate Lyase, CALCIUM ION, SULFATE ION | | Authors: | Motoshima, H, Iwatomo, Y, Watanabe, K, Oda, T, Muramatsu, T. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Alginate Lyase from Alteromonas sp.272
To be published
|
|
1UIH
 
 | | ANALYSIS OF THE STABILIZATION OF HEN LYSOZYME WITH THE HELIX DIPOLE AND CHARGED SIDE CHAINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Motoshima, H, Ohmura, T, Ueda, T, Imoto, T. | | Deposit date: | 1996-11-26 | | Release date: | 1997-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fluctuations in free or substrate-complexed lysozyme and a mutant of it detected on x-ray crystallography and comparison with those detected on NMR.
J.Biochem.(Tokyo), 131, 2002
|
|
1UIB
 
 | | ANALYSIS OF THE STABILIZATION OF HEN LYSOZYME WITH THE HELIX DIPOLE AND CHARGED SIDE CHAINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Motoshima, H, Ohmura, T, Ueda, T, Imoto, T. | | Deposit date: | 1996-11-26 | | Release date: | 1997-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fluctuations in free or substrate-complexed lysozyme and a mutant of it detected on x-ray crystallography and comparison with those detected on NMR.
J.Biochem.(Tokyo), 131, 2002
|
|
1UIF
 
 | |
1UID
 
 | |
1UIC
 
 | |
1UIE
 
 | |
1UIA
 
 | |
1UIG
 
 | |
8ZH8
 
 | | Human GPR103 -Gq complex bound to QRFP26 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2,Guanine nucleotide-binding protein G(i) subunit alpha-2,Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Iwama, A, Akasaka, H, Sano, F.K, Oshima, H.S, Shihoya, W, Nureki, O. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure and dynamics of the pyroglutamylated RF-amide peptide QRFP receptor GPR103.
Nat Commun, 15, 2024
|
|
2RU7
 
 | | Refined structure of RNA aptamer in complex with the partial binding peptide of prion protein | | Descriptor: | P16 peptide from Major prion protein, RNA_(5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3') | | Authors: | Hayashi, T, Oshima, H, Mashima, T, Nagata, T, Katahira, M, Kinoshita, M. | | Deposit date: | 2013-12-24 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Binding of an RNA aptamer and a partial peptide of a prion protein: crucial importance of water entropy in molecular recognition.
Nucleic Acids Res., 42, 2014
|
|
1KWK
 
 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase in complex with galactose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trimeric crystal structure of the glycoside hydrolase family 42 beta-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose.
J.Mol.Biol., 322, 2002
|
|
1KWG
 
 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trimeric Crystal Structure of the Glycoside Hydrolase Family 42 beta-Galactosidase from Thermus thermophilus A4 and the Structure of its Complex with Galactose
J.MOL.BIOL., 322, 2002
|
|
1J1R
 
 | | Structure of Pokeweed Antiviral Protein from Seeds (PAP-S1) Complexed with Adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Antiviral Protein S | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1J1S
 
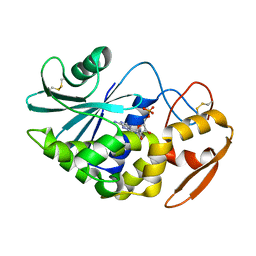 | | Pokeweed Antiviral Protein from Seeds (PAP-S1) Complexed with Formycin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiviral Protein S, FORMYCIN-5'-MONOPHOSPHATE | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1J1Q
 
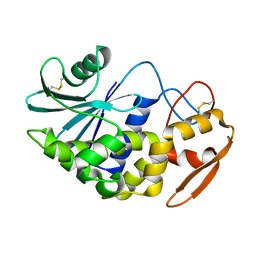 | | Structure of Pokeweed Antiviral Protein from Seeds (PAP-S1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiviral protein S | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
4PDT
 
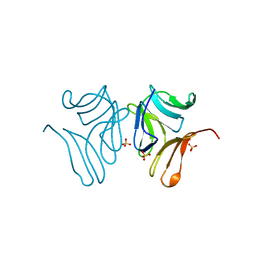 | | Japanese Marasmius oreades lectin | | Descriptor: | Mannose recognizing lectin, SULFATE ION | | Authors: | Noma, Y, Shimokawa, M, Maeganeku, C, Motoshima, H, Watanabe, K, Minami, Y, Yagi, F. | | Deposit date: | 2014-04-22 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of Japanese Marasmius oreades lectin at 1.40 Angstroms resolution.
To Be Published
|
|
4TKC
 
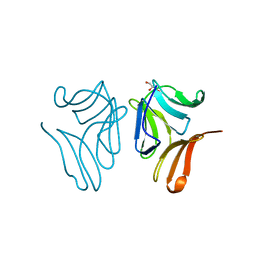 | | Japanese Marasmius oreades lectin complexed with mannose | | Descriptor: | GLYCEROL, Mannose recognizing lectin, alpha-D-mannopyranose, ... | | Authors: | Noma, Y, Shimokawa, M, Maeganeku, C, Motoshima, H, Watanabe, K, Minami, Y, Yagi, F. | | Deposit date: | 2014-05-26 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure of Japanese Marasmius oreades lectin complexed with mannose.
To Be Published
|
|
