1GSM
 
 | | A reassessment of the MAdCAM-1 structure and its role in integrin recognition. | | Descriptor: | MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1 | | Authors: | Dando, J, Wilkinson, K.W, Ortlepp, S, King, D.J, Brady, R.L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Reassessment of the Madcam-1 Structure and its Role in Integrin Recognition
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4AVA
 
 | | Crystal structure of protein lysine acetyltransferase Rv0998 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3FFZ
 
 | | Domain organization in Clostridium butulinum neurotoxin type E is unique: Its implication in faster translocation | | Descriptor: | ACETATE ION, Botulinum neurotoxin type E, SODIUM ION, ... | | Authors: | Kumaran, D, Eswaramoorthy, S, Swaminathan, S. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation.
J.Mol.Biol., 386, 2009
|
|
3GPK
 
 | |
3HDP
 
 | |
1Z0H
 
 | | N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Jayaraman, S, Eswarmoorthy, S, Ashraf, S.A, Smith, L.A, Swaminathan, S. | | Deposit date: | 2005-03-01 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
3CBN
 
 | |
7Z8N
 
 | | GacS histidine kinase from Pseudomonas aeruginosa | | Descriptor: | CALCIUM ION, Histidine kinase, R-1,2-PROPANEDIOL | | Authors: | Fadel, F, Bassim, V, Francis, V.I, Porter, S.L, Botzanowski, T, Legrand, P, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-03-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
4AVC
 
 | | Crystal structure of protein lysine acetyltransferase Rv0998 in complex with acetyl CoA and cAMP | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3H49
 
 | |
3B59
 
 | |
4CMV
 
 | | Crystal structure of Rv3378c | | Descriptor: | CITRIC ACID, DITERPENE SYNTHASE, MERCURY (II) ION | | Authors: | Layre, E, Lee, H.J, Young, D.C, Martinot, A.J, Buter, J, Minnaard, A.J, Annand, J.W, Fortune, S.M, Snider, B.B, Matsunaga, I, Rubin, E.J, Alber, T, Moody, D.B. | | Deposit date: | 2014-01-18 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Molecular Profiling of Mycobacterium Tuberculosis Identifies Tuberculosinyl Nucleoside Products of the Virulence-Associated Enzyme Rv3378C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3G1W
 
 | |
3IH0
 
 | | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized sugar kinase PH1459 | | Authors: | Kumar, G, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-29 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP
To be Published
|
|
3BE3
 
 | |
3HP0
 
 | |
4CMW
 
 | | Crystal structure of Rv3378c | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Layre, E, Lee, H.J, Young, D.C, Martinot, A.J, Buter, J, Minnaard, A.J, Annand, J.W, Fortune, S.M, Snider, B.B, Matsunaga, I, Rubin, E.J, Alber, T, Moody, D.B. | | Deposit date: | 2014-01-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Molecular Profiling of Mycobacterium Tuberculosis Identifies Tuberculosinyl Nucleoside Products of the Virulence-Associated Enzyme Rv3378C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3D0C
 
 | | Crystal structure of dihydrodipicolinate synthase from Oceanobacillus iheyensis at 1.9 A resolution | | Descriptor: | Dihydrodipicolinate synthase | | Authors: | Satyanarayana, L, Eswaramoorthy, S, Sauder, J.M, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-01 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase from Oceanobacillus iheyensis at 1.9 A resolution.
To be Published
|
|
4CMX
 
 | | Crystal structure of Rv3378c | | Descriptor: | 1,2-ETHANEDIOL, BENZENE HEXACARBOXYLIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Layre, E, Lee, H.J, Young, D.C, Martinot, A.J, Buter, J, Minnaard, A.J, Annand, J.W, Fortune, S.M, Snider, B.B, Matsunaga, I, Rubin, E.J, Alber, T, Moody, D.B. | | Deposit date: | 2014-01-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular Profiling of Mycobacterium Tuberculosis Identifies Tuberculosinyl Nucleoside Products of the Virulence-Associated Enzyme Rv3378C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3MMR
 
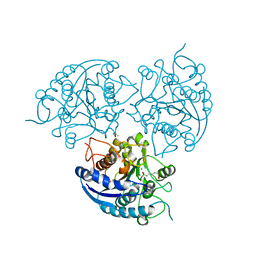 | | Structure of Plasmodium falciparum Arginase in complex with ABH | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase, BETA-MERCAPTOETHANOL, ... | | Authors: | Dowling, D.P, Ilies, M, Olszewski, K.L, Portugal, S, Mota, M.M, Llinas, M, Christianson, D.W. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of arginase from Plasmodium falciparum and implications for L-arginine depletion in malarial infection .
Biochemistry, 49, 2010
|
|
3DZB
 
 | |
3EWM
 
 | |
3DZ1
 
 | |
3EVZ
 
 | |
1SR7
 
 | | Progesterone Receptor Hormone Binding Domain with Bound Mometasone Furoate | | Descriptor: | GLYCEROL, MOMETASONE FUROATE, Progesterone receptor, ... | | Authors: | Madauss, K.P, Deng, S.-J, Austin, R.J, Lambert, M.H, McLay, I, Pritchard, J, Short, S.A, Stewart, E.L, Uings, I.J, Williams, S.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Progesterone receptor ligand binding pocket flexibility: crystal structures of the norethindrone and mometasone furoate complexes
J.Med.Chem., 47, 2004
|
|
