4D2S
 
 | | Human TTK in complex with a Dyrk1B inhibitor | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, N-{2-methoxy-4-[(1-methylpiperidin-4-yl)oxy]phenyl}-4-(1H-pyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine | | Authors: | Debreczeni, J.E, Kettle, J.G, Ballard, P, Bardelle, C, Butterworth, S, Colclough, N, Critchlow, S.E, Fairley, G, Fillery, S, Graham, M.A, Goodwin, L, Guichard, S, Hudson, K, Mahmood, A, Vincent, J, Ward, R.A, Whittaker, D. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Optimization of a Novel Series of Dyrk1B Kinase Inhibitors to Explore a Mek Resistance Hypothesis.
J.Med.Chem., 58, 2015
|
|
2OPS
 
 | | Crystal Structure of Y188C Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
4D2R
 
 | | Human IGF in complex with a Dyrk1B inhibitor | | Descriptor: | CHLORIDE ION, INSULIN-LIKE GROWTH FACTOR 1 RECEPTOR, N-{2-methoxy-4-[(1-methylpiperidin-4-yl)oxy]phenyl}-4-(1H-pyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine | | Authors: | Debreczeni, J.E, Kettle, J.G, Ballard, P, Bardelle, C, Butterworth, S, Colclough, N, Critchlow, S.E, Fairley, G, Fillery, S, Graham, M.A, Goodwin, L, Guichard, S, Hudson, K, Mahmood, A, Vincent, J, Ward, R.A, Whittaker, D. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of a Novel Series of Dyrk1B Kinase Inhibitors to Explore a Mek Resistance Hypothesis.
J.Med.Chem., 58, 2015
|
|
5AL9
 
 | | Structure of Leishmania major peroxidase D211R mutant (high res) | | Descriptor: | ASCORBATE PEROXIDASE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Chreifi, G, Hollingsworth, S.A, Li, H, Tripathi, S, Arce, A.P, Magana-Garcia, H.I, Poulos, T.L. | | Deposit date: | 2015-03-07 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Enzymatic Mechanism of Leishmania major Peroxidase and the Critical Role of Specific Ionic Interactions.
Biochemistry, 54, 2015
|
|
5DIS
 
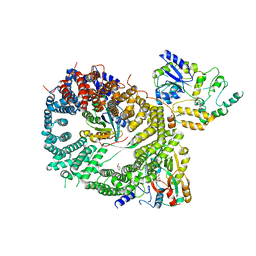 | | Crystal structure of a CRM1-RanGTP-SPN1 export complex bound to a 113 amino acid FG-repeat containing fragment of Nup214 | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Monecke, T, Port, S.A, Dickmanns, A, Kehlenbach, R.H, Ficner, R. | | Deposit date: | 2015-09-01 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Functional Characterization of CRM1-Nup214 Interactions Reveals Multiple FG-Binding Sites Involved in Nuclear Export.
Cell Rep, 13, 2015
|
|
1F31
 
 | |
3T4B
 
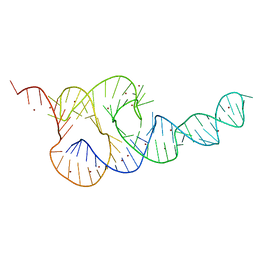 | | Crystal Structure of the HCV IRES pseudoknot domain | | Descriptor: | HCV IRES pseudoknot domain plus crystallization module, NICKEL (II) ION | | Authors: | Berry, K.E, Waghray, S, Mortimer, S.A, Bai, Y, Doudna, J.A. | | Deposit date: | 2011-07-25 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning.
Structure, 19, 2011
|
|
3SVL
 
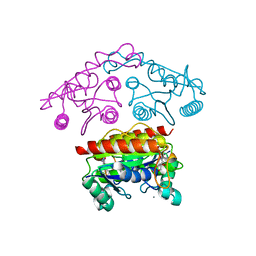 | | Structural basis of the improvement of ChrR - a multi-purpose enzyme | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, protein yieF | | Authors: | Poulain, S, Eswaramoorthy, S, Hienerwadel, R, Bremond, N, Sylvester, M.D, Zhang, Y.B, Van Der Lelie, D, Berthomieu, C, Matin, A.C. | | Deposit date: | 2011-07-12 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of ChrR-A Quinone Reductase with the Capacity to Reduce Chromate.
Plos One, 7, 2012
|
|
8H5V
 
 | | Crystal structure of the FleQ domain of Vibrio cholerae FlrA | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, flagellar regulatory protein A | | Authors: | Dasgupta, J, Chakraborty, S. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The N-terminal FleQ domain of the Vibrio cholerae flagellar master regulator FlrA plays pivotal structural roles in stabilizing its active state.
Febs Lett., 597, 2023
|
|
1C87
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 2-(OXALYL-AMINO-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Iversen, L.F, Andersen, H.S, Mortensen, S.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B.
J.Biol.Chem., 275, 2000
|
|
1C88
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Iversen, L.F, Andersen, H.S, Mortensen, S.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B.
J.Biol.Chem., 275, 2000
|
|
4AVB
 
 | | Crystal structure of protein lysine acetyltransferase Rv0998 in complex with acetyl CoA and cAMP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1B54
 
 | | CRYSTAL STRUCTURE OF A YEAST HYPOTHETICAL PROTEIN-A STRUCTURE FROM BNL'S HUMAN PROTEOME PROJECT | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, YEAST HYPOTHETICAL PROTEIN | | Authors: | Swaminathan, S, Eswaramoorthy, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-01-12 | | Release date: | 1999-01-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5AMM
 
 | | Structure of Leishmania major peroxidase D211N mutant | | Descriptor: | ASCORBATE PEROXIDASE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Chreifi, G, Fields, J.B, Hollingsworth, S.A, Heyden, M, Arce, A.P, Magana-Garcia, H.I, Poulos, T.L, Tobias, D.J. | | Deposit date: | 2015-03-11 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | "Bind and Crawl" Association Mechanism of Leishmania Major Peroxidase and Cytochrome C Revealed by Brownian and Molecular Dynamics Simulations.
Biochemistry, 54, 2015
|
|
1BJA
 
 | | ACTIVATION DOMAIN OF THE PHAGE T4 TRANSCRIPTION FACTOR MOTA | | Descriptor: | SULFATE ION, TRANSCRIPTION REGULATORY PROTEIN MOTA | | Authors: | Finnin, M.S, Cicero, M.P, Davies, C, Porter, S.J, White, S.W, Kreuzer, K.N. | | Deposit date: | 1998-06-23 | | Release date: | 1998-11-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The activation domain of the MotA transcription factor from bacteriophage T4.
EMBO J., 16, 1997
|
|
4CK8
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4-(4-(3,4- dichlorophenyl)piperazin-1-yl)phenylcarbamate (LFD) | | Descriptor: | (1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl {4-[4-(3,4-dichlorophenyl)piperazin-1-yl]phenyl}carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-30 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
4CKA
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (S)-1-(4-fluorophenyl)-2-(1H-imidazol-1-yl)ethyl 4- isopropylphenylcarbamate (LFS) | | Descriptor: | (1S)-1-(4-fluorophenyl)-2-(1H-imidazol-1-yl)ethyl 4-isopropylphenylcarbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
4CK9
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4- isopropylphenylcarbamate (LFT) | | Descriptor: | (1S)-1-(4-chlorophenyl)-2-(1H-imidazol-1-yl)ethyl [4-(propan-2-yl)phenyl]carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
3N2S
 
 | | Structure of NfrA1 nitroreductase from B. subtilis | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, NADPH-dependent nitro/flavin reductase | | Authors: | Morera, S, Gueguen-Chaignon, V, Meyer, P, Cortial, S, Ouazzani, J. | | Deposit date: | 2010-05-19 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NADH oxidase activity of Bacillus subtilis nitroreductase NfrA1: insight into its biological role.
Febs Lett., 584, 2010
|
|
1C86
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B (R47V,D48N) COMPLEXED WITH 2-(OXALYL-AMINO-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Iversen, L.F, Andersen, H.S, Mortensen, S.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B.
J.Biol.Chem., 275, 2000
|
|
1DIV
 
 | | RIBOSOMAL PROTEIN L9 | | Descriptor: | RIBOSOMAL PROTEIN L9 | | Authors: | Hoffman, D.W, Cameron, C, Davies, C, Gerchman, S.E, Kycia, J.H, Porter, S, Ramakrishnan, V, White, S.W. | | Deposit date: | 1996-07-02 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed RNA-binding protein.
EMBO J., 13, 1994
|
|
1EPW
 
 | |
3FKD
 
 | |
3U90
 
 | | apoferritin: complex with SDS | | Descriptor: | CADMIUM ION, DODECYL SULFATE, Ferritin light chain | | Authors: | Liu, R, Bu, W, Xi, J, Mortazavi, S.R, Cheung-Lau, J.C, Dmochowski, I.J, Loll, P.J. | | Deposit date: | 2011-10-17 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Beyond the detergent effect: a binding site for sodium dodecyl sulfate (SDS) in mammalian apoferritin.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3FCD
 
 | |
