5T49
 
 | | Crystal structure of SeMet derivative BhGH81 | | Descriptor: | 1,2-ETHANEDIOL, BH0236 protein, PHOSPHATE ION | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-08-29 | | Release date: | 2017-06-28 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of a Family 81 Glycoside Hydrolase Implicates Its Recognition of beta-1,3-Glucan Quaternary Structure.
Structure, 25, 2017
|
|
5T3B
 
 | | Crystal structure of BpGH50 | | Descriptor: | 1,2-ETHANEDIOL, Glycoside Hydrolase | | Authors: | Giles, K, Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-08-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a glycoside hydrolase family 50 enzyme from a subfamily that is enriched in human gut microbiome bacteroidetes.
Proteins, 85, 2017
|
|
5SUO
 
 | |
5ZK0
 
 | | Crystal structure of Peptidyl-tRNA hydrolase mutant -M71A from Vibrio cholerae | | Descriptor: | Peptidyl-tRNA hydrolase, SODIUM ION | | Authors: | Shahid, S, Kabra, A, Pal, R.K, Arora, A. | | Deposit date: | 2018-03-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Role of methionine 71 in substrate recognition and structural integrity of bacterial peptidyl-tRNA hydrolase.
Biochim. Biophys. Acta, 1866, 2018
|
|
6B1V
 
 | | Crystal structure of Ps i-CgsB C78S in complex with i-neocarratetraose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-09-19 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
5A29
 
 | | Family 2 Pectate Lyase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, EXOPOLYGALACTURONATE LYASE, MANGANESE (II) ION, ... | | Authors: | McLean, R, Hobbs, J.K, Suits, M.D, Tuomivaara, S, Jones, D, Boraston, A.B, Abbott, D.W. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional Analyses of Resurrected and Contemporary Enzymes Illuminate an Evolutionary Path for the Emergence of Exolysis in Polysaccharide Lyase Family 2.
J.Biol.Chem., 290, 2015
|
|
5T99
 
 | |
6B0K
 
 | | Crystal structure of Ps i-CgsB C78S in complex with k-carrapentaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-D-galactose, 4-O-sulfo-beta-D-galactopyranose, ... | | Authors: | Hettle, A, Boraston, A.B. | | Deposit date: | 2017-09-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
6B0J
 
 | | Crystal structure of Ps i-CgsB in complex with k-i-k-neocarrahexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2017-09-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Molecular Basis of Polysaccharide Sulfatase Activity and a Nomenclature for Catalytic Subsites in this Class of Enzyme.
Structure, 26, 2018
|
|
6FDZ
 
 | | Unc-51-Like Kinase 3 (ULK3) In Complex With Momelotinib | | Descriptor: | Momelotinib, Serine/threonine-protein kinase ULK3 | | Authors: | Mathea, S, Salah, E, Moroglu, M, Scorah, A, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Knapp, S. | | Deposit date: | 2017-12-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Unc-51-Like Kinase 3 (ULK3) In Complex With Momelotinib
To Be Published
|
|
5UE6
 
 | | Structure of nitrite reductase AniA from Neisseria gonorrhoeae, space group I4122 | | Descriptor: | COPPER (II) ION, Nitrite reductase, SODIUM ION | | Authors: | Hamza, A, Williamson, Z.A, Reed, R.W, Sikora, A.E, Korotkov, K.V. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Peptide Inhibitors Targeting the Neisseria gonorrhoeae Pivotal Anaerobic Respiration Factor AniA.
Antimicrob. Agents Chemother., 61, 2017
|
|
5A6J
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5ZHV
 
 | | Crystal structure of the PadR-family transcriptional regulator Rv3488 of Mycobacterium tuberculosis H37Rv in complex with zinc ion | | Descriptor: | Transcriptional regulator, ZINC ION | | Authors: | Meera, K, Pal, R.K, Arora, A, Biswal, B.K. | | Deposit date: | 2018-03-13 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of the transcriptional regulator Rv3488 ofMycobacterium tuberculosisH37Rv.
Biochem. J., 475, 2018
|
|
4JEB
 
 | | Crystal structure of an engineered RIDC1 tetramer with four interfacial disulfide bonds and four three-coordinate Zn(II) sites | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, ZINC ION, soluble cytochrome b562 | | Authors: | Tezcan, F.A, Medina-Morales, A.M, Perez, A, Brodin, J.D. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In Vitro and Cellular Self-Assembly of a Zn-Binding Protein Cryptand via Templated Disulfide Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4JE9
 
 | | Crystal structure of an engineered metal-free RIDC1 construct with four interfacial disulfide bonds | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562 | | Authors: | Tezcan, F.A, Medina-Morales, A.M, Perez, A, Brodin, J.D. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | In Vitro and Cellular Self-Assembly of a Zn-Binding Protein Cryptand via Templated Disulfide Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
5A55
 
 | | The native structure of GH101 from Streptococcus pneumoniae TIGR4 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-ALPHA-N-ACETYLGALACTOSAMINIDASE, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5AC5
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5AC4
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5A58
 
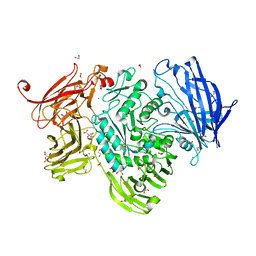 | | The structure of GH101 D764N mutant from Streptococcus pneumoniae TIGR4 in complex with serinyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A69
 
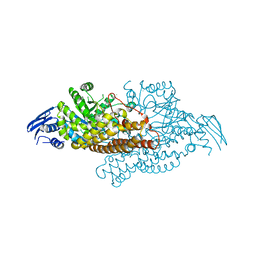 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with Gal-PUGNAc | | Descriptor: | N-ACETYL-BETA-D-GLUCOSAMINIDASE, [(Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5T98
 
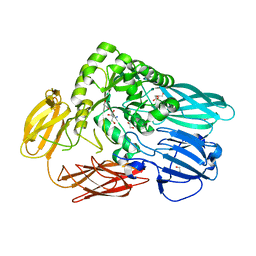 | | Crystal structure of BuGH2Awt | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside Hydrolase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5TA1
 
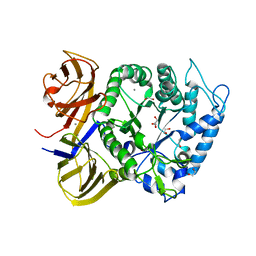 | | Crystal structure of BuGH86wt | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5TA9
 
 | |
4Z86
 
 | |
4ZXP
 
 | | Crystal structure of Peptidyl- tRNA Hydrolase from Vibrio cholerae | | Descriptor: | CITRATE ANION, Peptidyl-tRNA hydrolase | | Authors: | Shahid, S, Pal, R.K, Kabra, A, Yadav, R, Kumar, A, Arora, A. | | Deposit date: | 2015-05-20 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Unraveling the stereochemical and dynamic aspects of the catalytic site of bacterial peptidyl-tRNA hydrolase.
RNA, 23, 2017
|
|
